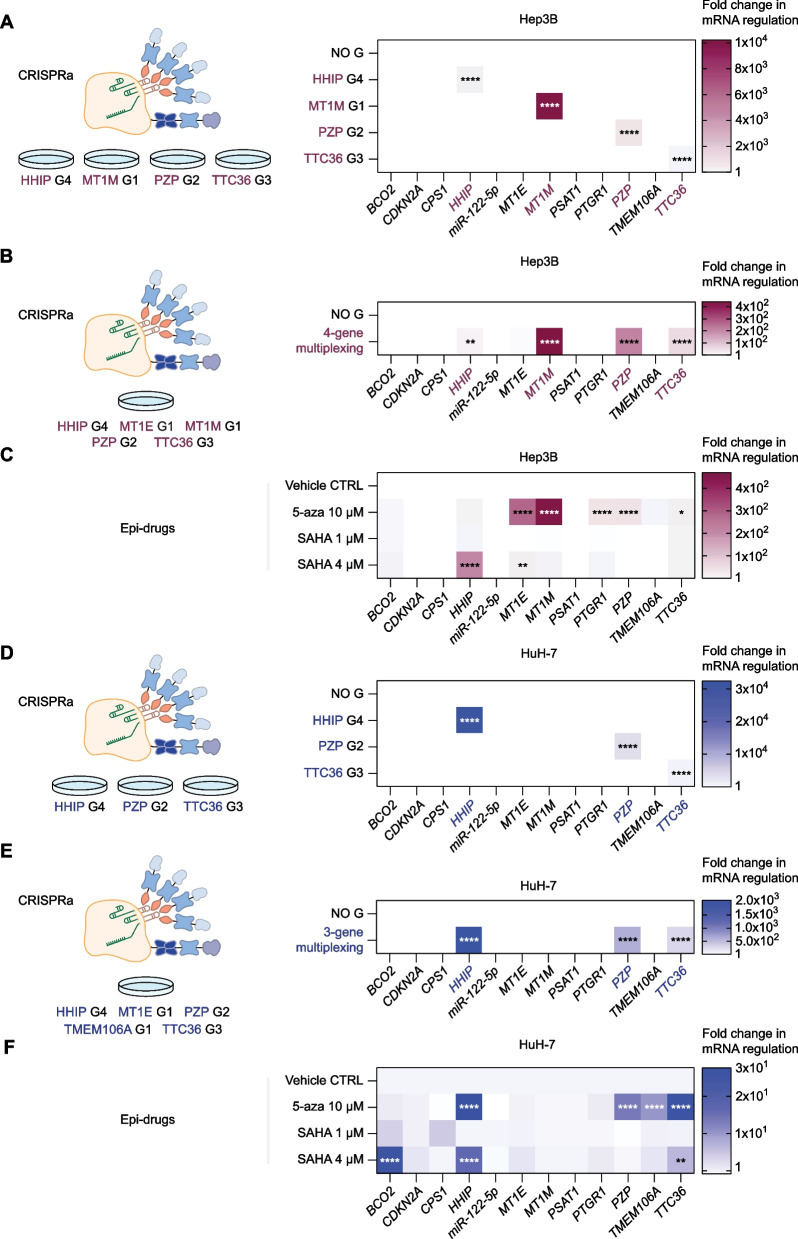
Fig. 6.
Specific and potent reactivation of silenced tumor suppressor genes by CRISPRa compared to epi-drugs, and CRISPRa-enabled genome multiplexing in Hep3B and HuH-7 HCC cells. A–F Heatmap of the 12-tumor suppressor gene (TSG) panel comparing the fold change in mRNA regulation evaluated by qRT-PCR 48 h after transient transfections (A), (B), (D) and (E), 72 h after 5-aza, or 48 h after SAHA treatments (C) and (F). The transfection conditions and the epi-drug treatments are arranged in rows, and the genes in columns. Hep3B and HuH-7 cells were either transfected with CRISPRa (SpdCas9-VPR and MS2-MCP-p65-HSF1) along with the most potent TSG-targeting gRNA, or with no gRNA (NO G) as control; or treated with epi-drugs or vehicle control. Relative gene expression was normalized to cells transfected with empty vector control and compared to CRISPRa with NO G (A), (B), (D), and (E), or normalized and compared to cells treated with vehicle control (C) and (F) for statistical analysis. Data presented as means (n = 3). A Targeted transcriptional regulation of four TSGs by CRISPRa in Hep3B cells. P values were determined by two-way ANOVA with Dunnett's multiple comparisons test (HHIP G4: ****P < 0.0001; MT1M G1: ****P < 0.0001; PZP G2: ****P < 0.0001; and TTC36 G3: **** P < 0.0001). B Targeted genome multiplexing for simultaneous transcriptional reactivation of four TSGs by CRISPRa in Hep3B cells. P values were determined by two-way ANOVA with Šídák's multiple comparisons test (HHIP: **P = 0.0092; MT1E: P = not significant; MT1M: ****P < 0.0001; PZP: ****P < 0.0001; and TTC36: ****P < 0.0001). C Untargeted transcriptional regulation of several TSGs in Hep3B cells treated with 10 µM 5-aza (MT1E: ****P < 0.0001; MT1M: ****P < 0.0001; PTGR1: ****P < 0.0001; PZP: ****P < 0.0001; and TTC36: *P = 0.0207), 1 µM SAHA, or 4 µM SAHA (HHIP: ****P < 0.0001; and MT1E: **P = 0.0089). P values were determined by two-way ANOVA with Dunnett's multiple comparisons test. D Targeted transcriptional regulation of three TSGs by CRISPRa in HuH-7 cells. P values were determined by two-way ANOVA with Šídák's multiple comparisons test. (HHIP G4: ****P < 0.0001; PZP G2: ****P < 0.0001; and TTC36 G3: ****P < 0.0001). E Targeted genome multiplexing for simultaneous transcriptional reactivation of three TSGs by CRISPRa in HuH-7 cells. P values were determined by two-way ANOVA with Šídák's multiple comparisons test (HHIP: ****P < 0.0001; MT1E: P = not significant; PZP: ****P < 0.0001; TMEM106A: P = not significant; and TTC36: ****P < 0.0001). F Untargeted transcriptional regulation of several TSGs in HuH-7 cells treated with 10 µM 5-aza (HHIP: ****P < 0.0001; PZP: ****P < 0.0001; TMEM106A: ****P < 0.0001; and TTC36: ****P < 0.0001), 1 µM SAHA, or 4 µM SAHA (BCO2: ****P < 0.0001; HHIP: ****P < 0.0001; and TTC36: **P = 0.0072). P values were determined by two-way ANOVA with Dunnett's multiple comparisons test