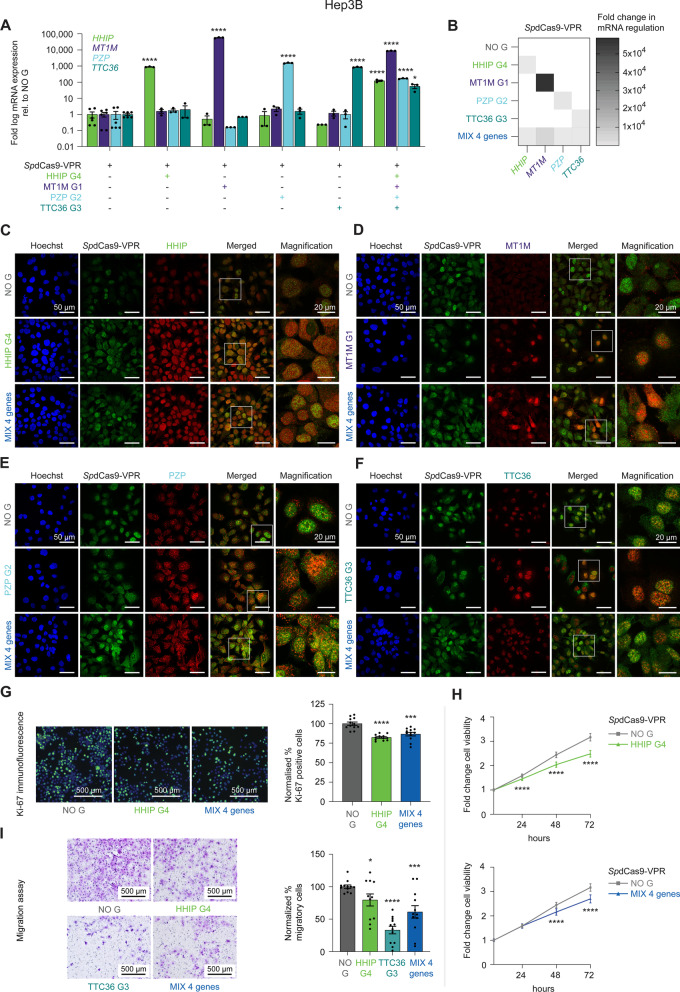
Fig. 7.
Reactivation of silenced tumor suppressor genes by CRISPRa correlates with phenotypic reprogramming in Hep3B HCC cells. A, B Transcriptional reactivation of four silenced tumor suppressor genes (TSGs), HHIP, MT1M, PZP, and TTC36, targeted individually and simultaneously (MIX 4 genes) by SpdCas9-VPR stably expressed along with the corresponding TSG-gRNA in Hep3B HCC cells. A Data shown as fold log10 change in TSG mRNA levels, evaluated by qRT-PCR, relative to SpdCas9-VPR with no gRNA (NO G). Data presented as means ± SEM (n = 3) and P values were determined by one-way ANOVA with Dunnett's multiple comparisons test (****P < 0.0001, *P = 0.0107). B Heatmap comparing the fold change in mRNA regulation evaluated by qRT-PCR. The TSG-gRNAs are arranged in rows, and the genes in columns. Data presented as means (n = 3). C–F Immunofluorescence of HHIP, MT1M, PZP, TTC36, SpdCas9-VPR, and Hoechst-stained cell-nuclei in stable Hep3B cells expressing SpdCas9-VPR alone (NO G), SpdCas9-VPR targeting HHIP with G4, MT1M with G1, PZP with G2, TTC36 with G3, or SpdCas9-VPR co-targeting all four TSGs (MIX 4 genes). G–I Phenotypic reprogramming in Hep3B cells lentivirally transduced with SpdCas9-VPR targeting and upregulating HHIP with G4, TTC36 with G3, the MIX 4 genes, or with NO G as control. G Cell proliferation assessed by α-Ki-67 immunostaining (green), superimposed on nuclear Hoechst 33258 staining (blue). Data normalized to SpdCas9-VPR NO G, presented as means ± SEM (n = 3), and P values were determined by unpaired t-test (****P < 0.0001, ***P = 0.0003). H Cell viability determined using a luminescence assay (CellTiter-Glo®). Data shown as fold change compared to SpdCas9-VPR NO G at 24, 48, and 72 h, presented as means ± SEM (n = 3), and P values were determined by unpaired t-test with Welch's correction (****P < 0.0001). I Inhibition of cell migration assessed by the Boyden chamber assay. Data normalized to SpdCas9-VPR NO G, presented as means ± SEM (n = 3), and P values were determined by unpaired t-test (*P = 0.0390, ****P < 0.0001, ***P = 0.0008)