Fig. 6.
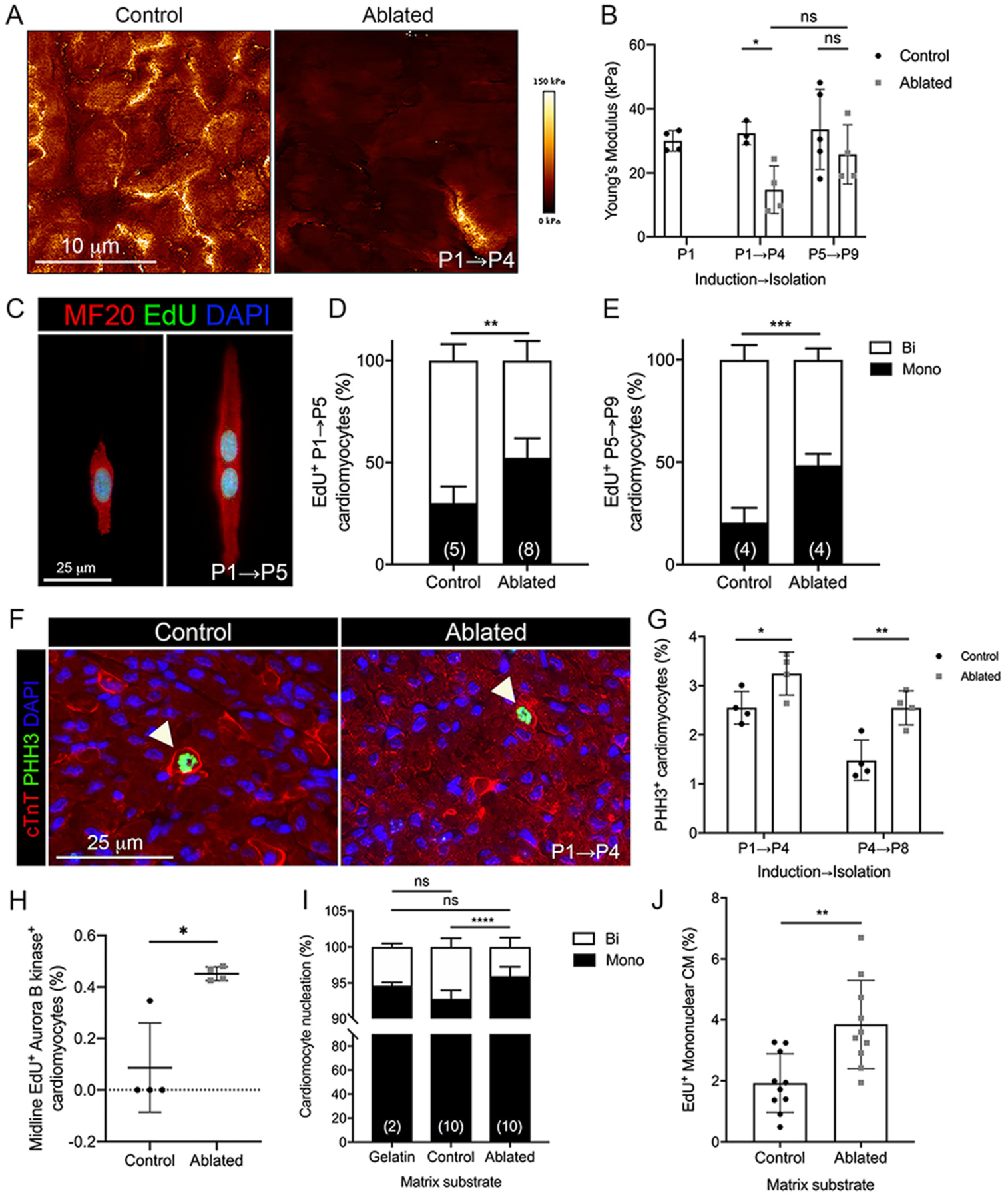
Cardiomyocyte cell cycle activity with fibroblast ablation. (A-B) Representative images of stiffness maps and the average Young’s modulus at indicated induction and isolation times. 4 scans of LV free wall per biological replicate were quantified. Control: n = 3–5; ablated: n = 4. (C) Representative images of EdU staining in isolated P5 cardiomyocytes. (D, E) Quantification of EdU+ mono and binucleated cardiomyocytes in (D) P5 and (E) P9 hearts induced at P1 and P5, respectively. Numbers within bar graphs represent biological replicate n values. 400–500 cardiomyocytes per biological replicate. (F) Representative images of immunostaining of cardiac troponin T (cTnT) and phosphohistone H3 (PHH3) in P4 heart. Arrows indicate PHH3+ cardiomyocytes. (G) Quantification of PHH3+ cardiomyocytes. n = 4 per group. 400–500 cardiomyocytes per biological replicate. (H) Quantification of midline EdU+/Aurora B kinase+ cardiomyocytes in P5 hearts. 200 cardiomyocytes per biological replicate. n = 4 per group. Quantification of (I) total mono and binucleated P2 cardiomyocytes and (J) EdU+ mononucleated P2 cardiomyocytes cultured on decellularized matrix from control and ablated P4 hearts for 48 h. Numbers within bar graphs represent biological replicate n values (control and ablated) or number of independent experiments (gelatin). 200 cardiomyocytes per biological replicate. Results are mean ± SD. Statistical significance was determined by unpaired t-test or Mann-Whitney U test (H). ns: not significant, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
