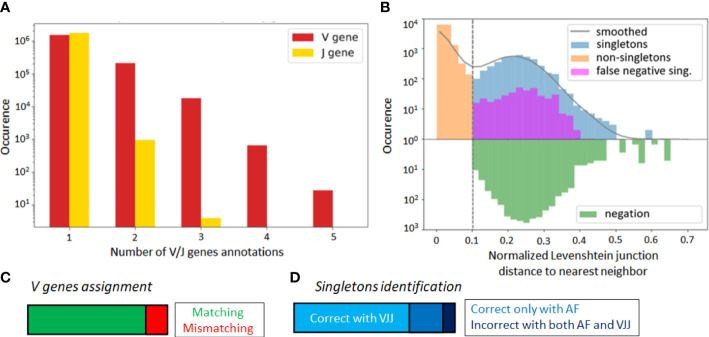
Figure 3.
(A) Occurrence rate of B-cell sequence with multiple V and J gene segment annotations of equivalent alignment score after the IgBlast query in the GC dataset. (B) Distance to nearest distribution for B-cell sequences in the alignment-based (VJ& Junction method) clonal identification method in the GC dataset. False negative singletons originating from V or J gene misalignment are depicted in purple. (C) Proportion of matched and mismatched V gene assignment between the original and truncated simulated dataset (where the first 70 nucleotides were artificially removed). (D) Correctly and wrongly assigned singletons by the VJJ and AF method on the simulated dataset.