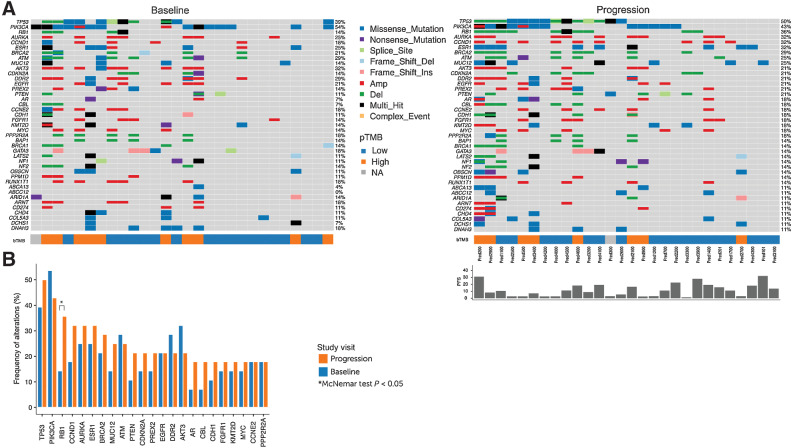
Figure 5.
Specific genomic alterations are enriched at disease progression. A, Heatmaps comparing the most frequently altered genes detected at progression versus baseline by PredicineWES+ (N = 28). Color coded bars below each heatmap indicate the level of bTMB, and the bar graph above the progression heatmap indicates PFS in months for each patient. Blue arrows denote patients with dominant APOBEC signatures. B, Enrichment at progression was observed for previously reported gene alterations implicated in resistance to endocrine and/or CDK4/6i therapy including a significantly higher frequency of RB1 alterations (P = 0.04; McNemar test), and nonsignificant enrichment in AR, AURKA, CCND1, CDKNKA, ESR1, FGFR, MYC, PTEN, and TP53 alterations. In addition, nonsignificant enrichment of gene alterations not commonly linked to ET + CDK4/6i resistance such as BRCA2, CBL, CDH, KMT2D, MUC12, and PREX2 was observed at progression. Two of the enriched gene alterations (MUC12 and PREX2) were not covered by the targeted PredicineATLAS panel and were detected only through comprehensive WES.