Diverse RNA-binding protein dependencies are identified for AML leukemic stem cells (LSC). Inhibition of ELAVL1 impairs splicing and RNA stabilization that supports LSC mitochondrial metabolism, identifying a novel therapeutic vulnerability.
Abstract
Acute myeloid leukemia (AML) is fueled by leukemic stem cells (LSC) whose determinants are challenging to discern from hematopoietic stem cells (HSC) or uncover by approaches focused on general cell properties. We have identified a set of RNA-binding proteins (RBP) selectively enriched in human AML LSCs. Using an in vivo two-step CRISPR-Cas9 screen to assay stem cell functionality, we found 32 RBPs essential for LSCs in MLL-AF9;NrasG12D AML. Loss-of-function approaches targeting key hit RBP ELAVL1 compromised LSC-driven in vivo leukemic reconstitution, and selectively depleted primitive malignant versus healthy cells. Integrative multiomics revealed differentiation, splicing, and mitochondrial metabolism as key features defining the leukemic ELAVL1–mRNA interactome with mitochondrial import protein, TOMM34, being a direct ELAVL1-stabilized target whose repression impairs AML propagation. Altogether, using a stem cell–adapted in vivo CRISPR screen, this work demonstrates pervasive reliance on RBPs as regulators of LSCs and highlights their potential as therapeutic targets in AML.
Significance:
LSC-targeted therapies remain a significant unmet need in AML. We developed a stem-cell–adapted in vivo CRISPR screen to identify key LSC drivers. We uncover widespread RNA-binding protein dependencies in LSCs, including ELAVL1, which we identify as a novel therapeutic vulnerability through its regulation of mitochondrial metabolism.
This article is highlighted in the In This Issue feature, p. 171
INTRODUCTION
Acute myeloid leukemia (AML) is a hematologic malignancy characterized by clonal expansion and accumulation of immature myeloid cells. To date, the effective treatment of AML remains a significant unmet clinical need, with standard-of-care underscored by dismal overall patient survival and high relapse rates (1). Decades of combined research in the mouse and human contexts have shown that populations of self-renewing leukemic stem cells (LSC) arising from mutations in hematopoietic stem or progenitor cells are both the seeds of initiation and the drivers of progression and relapse in AML (2, 3). Unlike bulk AML, LSCs exhibit unique cellular attributes including quiescence (4, 5), extensive self-renewal, distinct metabolism (6) and the capacity to localize to protective microenvironments (7, 8). Because of the critical role LSCs have in propagating AML, their effective and specific targeting represents a key therapeutic goal but one that is currently challenged by our poor understanding of the unique molecular drivers of the LSC state. Although in vivo CRISPR screening is gaining steam as a strategy to identify cancer and leukemia cell dependencies in general (9–11), to guide the discovery of the highest value therapeutic targets in AML, the uniqueness of the cancer stem cell state necessitates novel tailored high-throughput screening approaches that can a priori identify dependencies not just of progenitors and blasts, but of the LSCs themselves.
A focus on epigenetic and transcriptional changes that may underlie leukemic behavior has defined a large proportion of investigations into AML-specific targets to date; however, the extensive posttranscriptional layer as it pertains to LSC function has received comparably little attention. Here, RNA-binding proteins (RBP) are core effectors, rapidly executing precise control of gene expression by modulating a diversity of RNA properties that include splicing, polyadenylation, localization, stabilization, degradation, and translation. Through the association of their RNA-binding domains (RBD) with consensus sequences in their targets, each RBP can link the fate of many, often functionally related, mRNAs (12). When dysregulated, RBPs can contribute to disease pathology, as one genomic study revealed that 50% of RBPs are mutated across a variety of cancer types (13), and in AML there exist isolated examples of RBPs that have been uncovered as specific pro-LSC factors (14–17) notwithstanding, in some cases, their necessity for normal HSCs (18–21). Despite these intriguing cases and the emerging evidence of the importance of RBP-mediated posttranscriptional control in cancer progression (22–26), this level of regulation has not been systematically explored in AML LSCs. Moreover, with the complement of RBPs on the order of 2,000, these regulators represent a potentially enormous untapped source for therapeutic target discovery.
In addressing these questions, herein we identified a large subset of RBPs selectively enriched in the LSC fraction of AML by interrogating stem and progenitor cell populations in healthy bone marrow (BM) and AML patient samples. We devised a unique two-step serial transplantation in vivo CRISPR-Cas9–mediated pooled dropout screen that identified 32 RBPs underlying LSC function and thus of elevated translational value. These targets, which span a diverse set of RBPs, include the RNA stabilizing factor ELAVL1. We demonstrate the therapeutic potential of targeting LSCs while sparing healthy stem cell counterparts via small-molecule inhibition of ELAVL1 in patient-derived AML xenografts, and through comprehensive multiomics profiling of its posttranscriptional regulon reveal mitochondrial metabolism and the mitochondrial protein import regulator, TOMM34, as a key axis through which ELAVL1 sustains LSC function and AML survival.
RESULTS
Identification of an RBP Subset Uniquely Enriched in LSCs
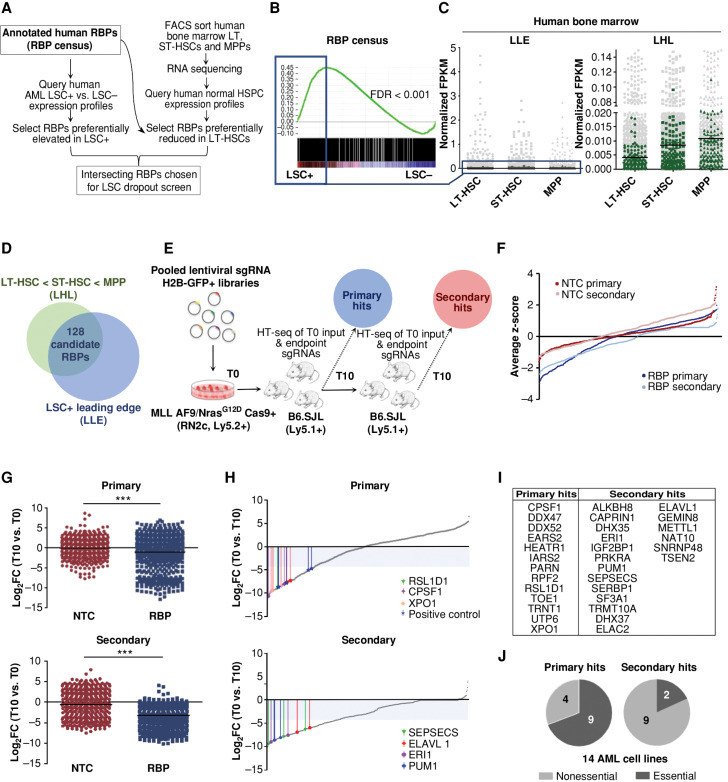
To identify a set of RBPs with potential roles in the selective control of human LSCs, we performed an expression study of genes encoding proteins encompassed within the human RBP census (27). Interestingly, as a class RBPs were revealed as significantly enriched in LSCs by gene set enrichment analysis (GSEA) of a data set of transcriptionally profiled and functionally validated LSC+ and LSC− fractions obtained from 78 AML patients (ref. 28; Fig. 1A and B). Within this GSEA, we defined the top 500 LSC-enriched RBPs (blue box in Fig. 1B) as LSC leading-edge (LLE) and evaluated their expression profiles in the following primitive fractions of human BM: long-term (LT-) HSC, short-term (ST-) HSC and multipotent progenitors (MPP; Supplementary Table S1; Fig. 1A and C). Overall, LLE RBPs were expressed at much lower levels across these BM subpopulations compared with the entire RBP census (Fig. 1C, left). Additionally, we identified a subset of RBPs within the LLE that exhibit uniquely low and/or relatively reduced expression in the LT-HSC compartment relative to more committed ST-HSCs and MPPs, which we termed LT-HSC low (LHL) RBPs (Fig. 1C, right). Upon intersecting the LHL and LLE RBPs, we identified a group of 128 RBPs elevated in LSCs compared with HSPCs and especially lower expression in the LT-HSC (Fig. 1A and D). Because of their low expression in normal LT-HSCs, indicating possibly reduced importance for normal hematopoietic function, we nominated the 128 RBP subset as potentially critical selective LSC determinants with high therapeutic relevance.
Figure 1.
Diverse RBPs are enriched in LSCs and identified as in vivo AML LSC essentialities through a two-step pooled in vivo CRISPR-Cas9 dropout screen. A, Overview of in silico selection of RBPs preferentially heightened in LSCs and reduced in LT-HSCs. B, GSEA plot showing LSC-enriched RBPs. The top 500 of the leading-edge LSC-enriched RBPs (LLE) are indicated by the blue box. C, Expression of LSC-enriched RBPs (LLE) in LT-HSC, ST-HSC, and MPP populations of human BM (dark gray, left) relative to all RBPs in census (light gray, left) and a subset of RBPs (LHL) with lowest expression in LT-HSCs of human BM (green, right) relative to all RBPs in census (light gray, right). D, Selection of the 128 RBPs exhibiting LSC-enriched and LT-HSC-reduced expression. E, Schematic illustrating the in vivo dropout screen. HT-seq, high-throughput sequencing. F, Average ranked dropout z-scores for all sgRNAs in both arms and transplantation rounds. G, Median log2 fold-change (T10 vs. T0) of unique sgRNAs in the NTC and RBP arms of the screen at the primary (top) and secondary (bottom) endpoints are shown. H, Median LFC of all sgRNAs within the RBP arm of the screen after the primary (top) and secondary (bottom) rounds. Select top-scoring sgRNAs are indicated with colored bars; shaded area indicates the decreased fold change of 20 cutoff. I, RBPs are called hits across primary and secondary screening arms. J, Analysis of general essentiality across a panel of AML cell lines (40) for the RBPs considered hits at primary endpoints (primary, left) compared with RBP hits where all targeting sgRNAs dropped out only in secondary recipients (secondary, right). A gene was considered generally essential if its average log2 fold change in abundance was less than −1 in at least 12 of the 14 tested AML lines in vitro. n = 2–3 mice per 1° T10 and 2° T0 and T10 replicates. LT-HSC = CD34+CD38−CD90+CD49f+; ST-HSC = CD34+CD38−CD90+CD49f−; MPP = CD34+CD38−CD90−CD49f−. ***, P < 0.001, determined by a two-sided Student t test.
A Pooled Two-Step In Vivo RBP CRISPR Dropout Screen in MLL-AF9/NrasG12D Leukemia
Primary LSCs are rare (29), and once isolated, rapidly lost in culture. Consequently, in vitro leukemia screens predominately identify genes essential for proliferation (a characteristic property of progenitor cells) but fail to pinpoint genes required for repopulation or self-renewal (hallmark features of bona fide stem cells). Therefore, true LSC function can only be assayed in vivo by evaluating the capacity of cells to regenerate serially transplantable leukemia (29, 30). To this point we designed a two-step in vivo pooled CRISPR-Cas9 screening approach that would identify candidates within the 128 LSC-enriched RBPs capable of regulating the functional property of leukemia reconstitution as measured by primary transplantation. As heightened self-renewal of LSCs contributes to their capacity to serially propagate leukemia, we reasoned that a secondary transplantation step would identify RBPs that uniquely control LSC self-renewal (Fig. 1E). We selected a Cas9-expressing MLL-AF9/NRasG12D mouse leukemia (RN2c) as our in vivo system (31), on the basis of its immunophenotypically well-defined and relatively abundant LSC fraction, validated surrogacy of human MLL-AF9 counterpart disease, including its therapeutic targets and its heightened expression of the candidate RBPs to be screened as compared with healthy mouse hematopoietic stem and progenitor cells (HSPC; refs. 30, 32–35; Supplementary Fig. S1A). We designed lentiviral sgRNA constructs (31, 36) targeting all 128 LSC-enriched RBPs (4–5 sgRNAs per RBP) wherein annotated early exon RNA-binding motifs were targeted where possible to encourage maximum negative selection. Positive control sgRNAs that impair in vivo RN2c cell fitness (31) were combined with >400 nontargeting control (NTC) sgRNAs in a separate arm of the screen (Supplementary Table S2). RN2c cells were infected with either the NTC or RBP-targeting library pools and serially transplanted into recipient mice, with sgRNA representation captured postinfection and at each transplant endpoint (Fig. 1E).
Over the course of transplantation, transduced H2B-GFP+ fractions in the RBP-targeting arm gradually decreased, whereas in the NTC arm, the H2B-GFP levels remained stable (Supplementary Fig. S1B), findings paralleled at the level of sgRNA representation (Fig. 1F; Supplementary Fig. S1C and S1D). When quantitatively assessed using the MAGeCK algorithm (37), the median log2 fold-change (LFC) of sgRNA abundance as compared with that on the day of transplant was significantly lower at the end of the primary transplant compared with that on the day of transplant in the RBP arm, but not in the NTC arm, and this further increased in magnitude following secondary screening, indicating selective and progressive loss of RBP-targeting sgRNAs (Fig. 1G; Supplementary Fig. S1E).
Classification of Primary and Secondary Depleting sgRNAs and Target RBPs
To identify RBPs important for leukemic repopulation and LSC-driven propagation, we set a stringent threshold where at least 2 of its targeting sgRNAs must drop out with a depletion greater than an LFC of −4, a threshold reached for all positive controls tested (Supplementary Fig. S1F). Using this selection criterion, we identified 32 hit RBPs, with 13 RBPs reaching our threshold in primary recipients (“primary hits”) and 19 achieving the 2 sgRNA depletion threshold only upon passage through secondary recipients (“secondary hits”; Fig. 1H and I). GO annotations indicate that the screen hits are involved in diverse RNA metabolic processes/interactions and molecular pathways including nitrogen compound metabolism, tRNA processing, ribosome biogenesis, and mRNA processing, indicating dependence on a broad range of posttranscriptional regulation (Supplementary Fig. S1G and SH). The most significant hit RBPs for which the greatest number of sgRNAs dropped out include RSL1D1, CPSF1 (a mRNA cleavage and polyadenylation factor), and XPO1 (mediator of RNA nuclear export) in primary transplants, and ELAVL1, SEPSECS1, ERI1, and PUM1 as significant dropouts only in the secondary round, each distinguished by diverse roles in RNA control with functions ranging from mRNA stabilization, tRNA, selenocysteine synthesis, histone mRNA degradation and translational repression, respectively (Fig. 1H). Of note, XPO1 and PUM1 have been identified as drivers in human AML LSCs and mouse leukemia, respectively (38–40), supporting our screen's capacity to identify bona fide LSC regulators.
We next analyzed all screen hit RBPs against a previously reported Gene Essentiality analysis of genome-scale in vitro CRISPR-Cas9 screening of a panel of 14 AML cell lines. Notably, 69.2% (9/13) of primary hits were found to be generally critical for leukemia cell line propagation whereas only 42.0% (8/19) secondary hit RBPs were found to be critical for growth across the majority of lines (41). Importantly when considering those secondary hits in which all sgRNA dropouts called occurred only within secondary recipients, essentiality in vitro was found for only 18.2% (2/11; Fig. 1J; Supplementary Fig. S1I). Altogether, our results suggest that serial in vivo screening can effectively identify LSC drivers including genes regulating LSC repopulation and self-renewal that appear underrepresented or dispensable in in vitro dependency screens. Moreover, our results showcase the novel insights that can be gained using this two-step in vivo screening approach where the unique secondary screening arm we have used served to uncover a host of effects masked in the primary transplant arm. Given the critical contribution of LSC self-renewal in serial reconstitution, this secondary screening arm thus has a high likelihood of having captured bona fide LSC-specific events.
Validation of Individual sgRNAs Identifies ELAVL1 as a Top-Scoring RBP
To validate the outcome of our two-step in vivo drop out screen, we selected top-scoring primary and secondary hit RBPs for individual knockout in RN2c and following their independent repression we assessed the effects in vitro, compared the in vivo growth dynamics to that observed in the screen, and quantified the LSC compartment within the leukemic grafts. For each sgRNA used, we verified highly efficient CRISPR-induced indel formation at the targeted locus (ref. 42; Supplementary Fig. S2A). Next, we evaluated in vivo propagation of RN2c transduced with these individual sgRNAs in comparison with a nonessential control sgRNA targeting Ano9 (ref. 31; Fig. 2A). In line with the screen results, we observed that sgRNAs targeting the primary hits (CPSF1 and RSL1D1) strongly depleted in the first round of transplantation, whereas sgRNAs targeting secondary hits (ELAVL1, SEPSECS, and ERI1) showed more moderate depletion of H2B-GFP+ cells in the primary transplantation followed by a strong depletion upon secondary transplantation (Fig. 2B and C). These results confirmed that individual knockout events replicated their dynamics as tracked in the pooled screen setting. We next assessed the effects of these sgRNAs in cultured RN2c cells and found that in all cases the knockout of our hit RBPs exhibited either negligible or less detrimental effects on in vitro growth in comparison with those observed in the long-term in vivo context (Supplementary Fig. S2B). Lastly, we quantified the effect of individual gene depletions on the LSC-enriched cKit+ fraction (32, 35) within detectable H2B-GFP+ grafts. We observed a relative decrease of this fraction for all sgRNAs tested supporting an LSC-specific impairment of RN2c cells in vivo upon genetic ablation of hit RBPs (Fig. 2D). The approach of considering both canonical and noncanonical RBPs, selecting candidates based on selectively elevated expression patterns in LSC and pairing this with screening in an in vivo serial transplantation setting thus allowed us to uncover unique regulators of LSCs highlighting the potential functional and clinical relevance of RBPs in AML.
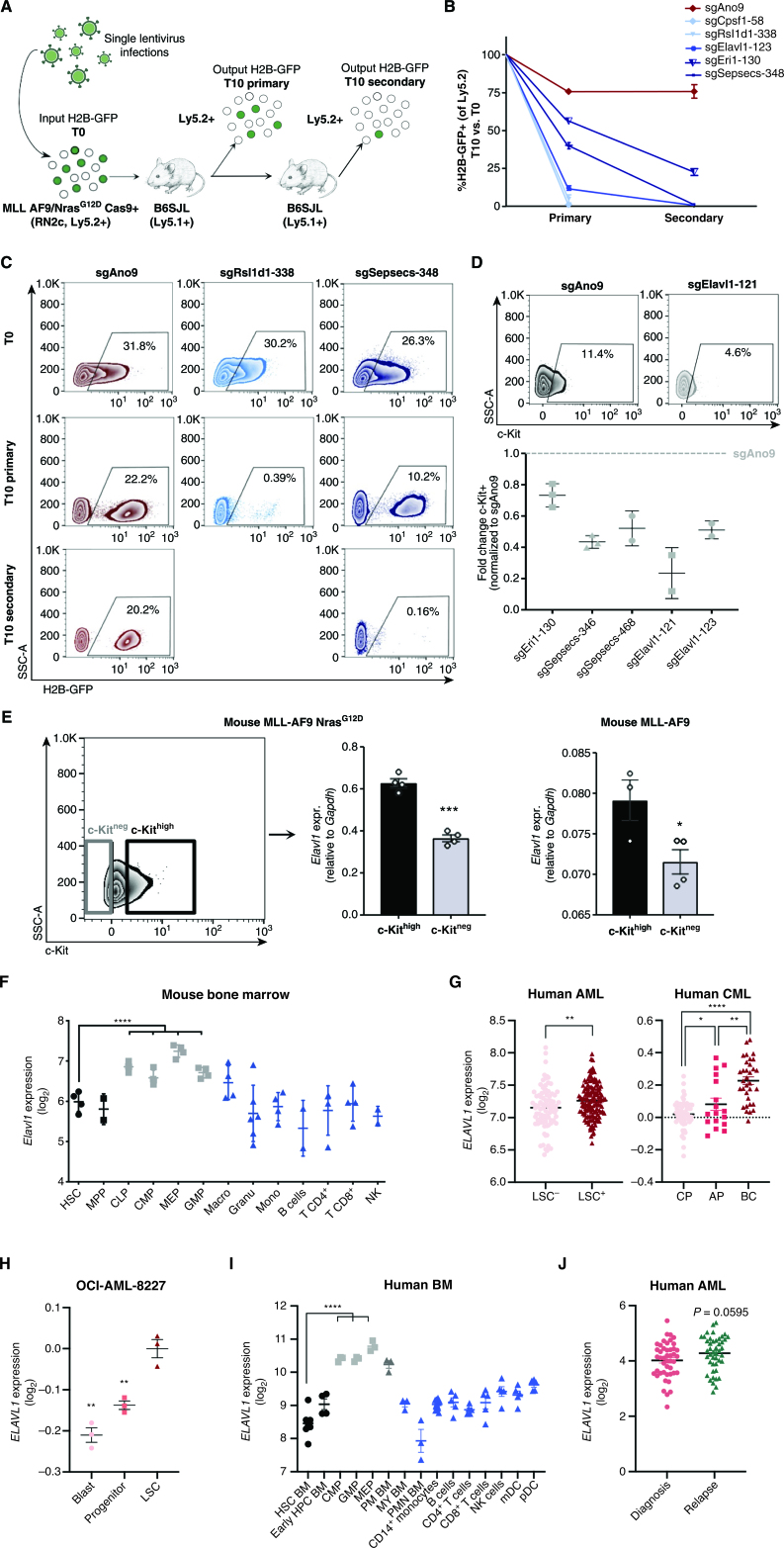
Figure 2.
Independent hit knockout validation replicates pooled CRISPR-Cas9 screen dropout dynamics and identifies ELAVL1 as a top LSC dependency. A, Schematic illustrating the in vivo screen validation strategy. B, Percentage of H2B-GFP+ cells in the output graft as compared with the T0 input. sgAno9 is the negative control and darkening shades of blue correlate with increasing time in vivo before sgRNA dropout. C, Representative flow plots of RN2c cells sampled at each time point (T0, T10 primary, T10 secondary) are shown. D, Fold change (FC) of cKit+ fractions within H2B-GFP+ populations of BM samples with >5% H2B-GFP+ of Ly5.2+. E, Transcript expression of Elavl1 in LSC+ (cKithigh, top 25%) and LSC− (cKitneg) MLL-AF9 NrasG12D (RN2c) cells (left; flow sorting gates are shown) and MLL-AF9 cells (right). F, Levels of Elavl1 mRNA across various subpopulations of the mouse hematopoietic hierarchy, adapted from BloodSpot. Mature hematopoietic lineages, progenitor populations, and primitive populations are shown in blue, gray, and black, respectively. G, Human ELAVL1 expression in LSC+ and LSC− AML subfractions (27) and throughout CML disease stages (adapted from ref. 46 and www.oncomine.org) are shown in left and right, respectively. H, Expression of ELAVL1 in subpopulations of OCI-AML-8227 cell line. I, Transcript levels of ELAVL1 across sorted subfractions of the human BM hematopoietic hierarchy, adapted from BloodSpot (43). Mature hematopoietic lineages, progenitor populations, and primitive populations are shown in blue, gray, and black, respectively. J,ELAVL1 expression levels across paired diagnosis-relapse primary human AML samples. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, as determined by a two-sided Student t test. Error bars, SEM. AP, accelerated phase; CP, chronic phase.
From all of the top-scoring secondary hit RBPs independently validated, the sgRNA with the strongest depletion of cKit+ cells over the course of in vivo leukemic propagation was one that targets Elav-like protein 1 (Elavl1), an RBP primarily characterized for its role in regulating gene expression via stabilization of its RNA targets (Fig. 2D). Additionally, whereas deletion of ELAVL1 resulted in pronounced in vivo depletion of RN2c, a more moderate growth inhibition was observed in vitro (Supplementary Fig. S2B). Likewise, in the human AML cell line THP-1, which also possesses MLL-AF9/NRasG12D mutations, we found that CRISPR-mediated knockout of ELAVL1 had very low magnitude effects on cell death and yielded only a modest ∼20% reduction in cell growth and progenitor CFU output (Supplementary Fig. S2C–S2G). Furthermore, using shRNA-mediated depletion of ELAVL1 in four individual AML cell lines, we observed extremely low overall cell death (<3% Annexin V+7AAD+ cells) in the THP-1, MOLM-13, and MV-411 lines, despite having increased cell death as compared with control (Supplementary Fig. S2H). Conversely, ELAVL1-depleted NOMO-1 cells showed significantly lower cell death in comparison with their shScramble controls, but still maintained <6% cell death across all shRNA (Supplementary Fig. S2H). Altogether, this implicates ELAVL1 as an LSC regulator that may be underprioritized by an in vitro screening approach. Furthermore, we found that the LSC-enriched cKithigh expressing fraction of RN2c cells showed significantly elevated transcript levels of Elavl1 compared with cKitneg cells (Fig. 2E, left and middle). This was also observed in the cKithigh and cKitlow fractions of a separately derived MLL-AF9 mouse model devoid of Ras mutations (Fig. 2E, right, Supplementary Fig. S2I–S2K), indicating an LSC-specific increased expression of this RBP in MLL-AF9–driven leukemias. We next examined the expression of Elavl1 in publicly available data sets of mouse HSPCs (43, 44). Compared with MPPs, ELAVL1 is decreased at the protein level in the HSC fraction of normal mouse BM (Supplementary Fig. S2L). Moreover, Elavl1 shows a progressive elevation in expression within mouse HSPCs peaking in downstream megakaryocyte-erythroid progenitors (MEP; Fig. 2F), indeed suggesting that its heightened expression in the stem cell context could be unique to AML.
Human and mouse ELAVL1 show >90% protein sequence conservation (45), and consistent with having passed the expression filters necessitated by the screen's candidate selection strategy, ELAVL1 transcripts are significantly enriched in human AML LSC+ fractions (ref. 28; Fig. 2G, left). When analyzed in a data set of 91 BCR-ABL–driven chronic myeloid leukemia (CML) patient samples, a gradually increasing pattern is observed for ELAVL1 transcript levels across the progressively more aggressive chronic to blast crisis (BC) phases with the highest levels observed in BC (Fig. 2G, right), the most LSC-enriched phase (46). Additionally, the expression of ELAVL1 in a primary AML model system propagated in vitro but possessing a defined functional hierarchy (OCI-AML-8227) demonstrates a significant enrichment in the most primitive population of LSCs compared with downstream clonogenic progenitors and terminal blasts (ref. 47; Fig. 2H). Importantly, consistent with the mouse system, ELAVL1 levels are lower in human HSCs and show a step-wise increase throughout the human HSPC hierarchy peaking in MEPs (Fig. 2I). Moreover, when ELAVL1 is evaluated in the bulk cells across a cohort of primary AML samples representing 27 distinct categories of cytogenetic abnormalities, it is in almost every case heightened in expression in leukemic blasts relative to normal HSC counterparts (refs. 48–53; Supplementary Fig. S2M), indicating that its elevation in leukemia is common and largely agnostic to underlying genetic abnormalities. Above median levels of ELAVL1 expression did not show any significant prognostic trend in two independent cohorts of AML patients (53, 54); however, in a pooled set of expression profiles from 39 paired AML diagnosis-relapse samples (3, 55, 56), ELAVL1 expression is significantly increased upon relapse (Fig. 2J). Our in vivo screening results considered together with these robust expression profiles in normal and leukemic samples are strongly predictive that ELAVL1 could be an important cross-cutting driver of LSC function across leukemic subtypes with potentially reduced dependence in healthy HSCs.
Depletion of ELAVL1 Expression Selectively Impairs the Murine LSC Compartment
Given the potentially selective importance of ELAVL1 in LSCs, we next took an RNAi-based approach to pursue loss-of-function studies in multiple genetically diverse LSCs and normal HSPCs. Using our lentiviral systems, we knocked down ELAVL1 in both non-Ras mutated MLL-AF9 and bcCML mouse leukemias (Fig. 3A; Supplementary Table S2). We observed significantly decreased colony-forming ability in shElavl1-transduced MLL-AF9 cells relative to shLuciferase controls (Fig. 3B; Supplementary Fig. S3A). Upon serial transplantation of infected MLL-AF9 BM cells, we observed a significant loss of transduced Ametrine+ populations within shElavl1-transduced secondary grafts suggesting that leukemic cells, and LSCs in particular, are sensitive to reduced ELAVL1 (Fig. 3C). Furthermore, cell-surface marker analysis revealed that, in contrast to controls, the LSC-enriched cKithigh fraction (33, 34) of shElavl1-infected populations specifically decreased over the course of in vivo propagation (Fig. 3D). When repeated in the distinct LSC-driven bcCML mouse model that has been used to dissect clinically relevant insights into the corresponding human disease (Supplementary Fig. S3B–S3E; refs. 57–59), we again observed impaired serial in vivo leukemic reconstitution upon knockdown of ELAVL1 (Supplementary Fig. S3F). Moreover, shLuciferase- and shElavl1-transduced bcCML LSCs revealed increased levels of apoptosis upon ELAVL1 knockdown (Supplementary Fig. S3G and S3H). These findings demonstrate that in genetically distinct types of mouse myeloid leukemia, ELAVL1 repression has significant inhibitory effects on LSC-mediated in vivo leukemic growth.
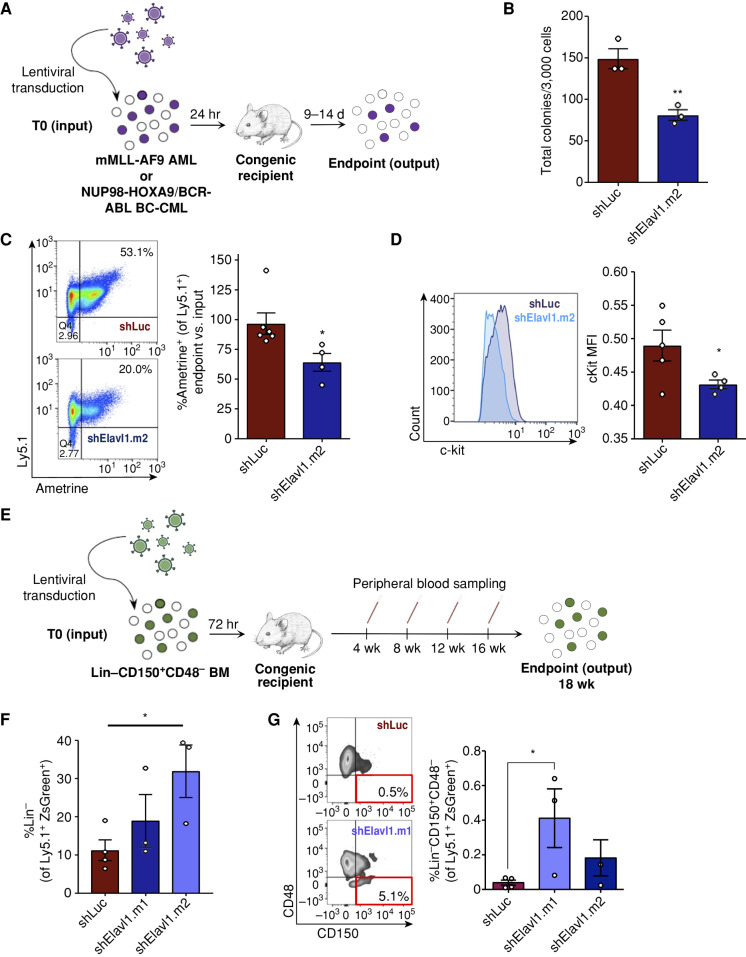
Figure 3.
Elavl1 knockdown impairs in vivo leukemic propagation and spares healthy LT-HSCs. A, Schematic illustrating ELAVL1 loss-of-function in vivo transplantation assays in mMLL-AF9 and bcCML mouse models. B, CFU output from shLuciferase- and shElavl1-infected MLL-AF9 mouse leukemic BM, 10 days after plating (n = 3). C and D, Flow-cytometric analysis of the normalized output vs. input Ametrine+ fractions (C) and c-Kit mean fluorescence intensity (D) of shLuciferase- and shElavl1-infected MLL-AF9 BM at the secondary transplant endpoint. Representative flow plots and histograms are shown on the left in each panel. E, Schematic of in vivo evaluation of ELAVL1 knockdown in normal mouse BM stem and progenitor cells. F and G, Flow-cytometric analysis of Ly5.1+ZsGreen+ BM showing Lin− (F) and Lin−CD150+CD48− (G) fractions at the 18-week after transplant endpoint. *, P < 0.05, determined by a two-sided Student t test. Error bars, SEM.
We next wanted to assess the effect of a similar level of ELAVL1 repression on normal stem cell function, as ideal antileukemic therapeutics should spare healthy HSCs. Previous reports present contrasting conclusions on the effects of ELAVL1 loss in primitive murine hematopoietic cells. In a conditional knockout model, inducible deletion of ELAVL1 in native mice did not alter BM Lin−Sca-1+c-Kit+ (LSK) percentages on short-term follow-up (60), whereas a separate study suggested LSK Flt3+CD34− cells are reduced in the 12-week grafts derived from primitive BM cells transduced with a single hairpin directed against Elavl1 (61). Methodological differences and the possibility for off-target effects in the shRNA study complicate the interpretation of these disparate findings. In addition, Yilmaz and colleagues have reported that inclusion of SLAM family markers, which were not explored in the latter study, markedly increase the purity of HSCs from reconstituted mice (62). To address this, we lentivirally delivered shRNAs (ZsGreen+) into Lin−CD150+CD48− mouse HSCs (Ly5.1+) and assayed their function over long-term hematopoiesis by competitive transplantation (Fig. 3E). At 4 weeks after transplant, during which early multipotent progenitors are the dominant contributors to reconstitution levels, the shElavl1-infected grafts in the peripheral blood showed a significant decrease of ∼40% compared with control. After this point, however, changes to ZsGreen+ proportions in shElavl1 grafts normalized and tracked closely to control grafts (Supplementary Fig. S3I). Most importantly, at the 18-week endpoint, the mean percentage of Lin− and the highly HSC-enriched Lin−CD150+CD48− fraction within the shElavl1 BM grafts were either elevated or not reduced relative to shLuciferase (Fig. 3F and G). In addition, we observed no lineage skewing within shElavl1 grafts (Supplementary Fig. S3J). Together, our findings suggest that although constitutive repression of ELAVL1 impairs an early population of progenitors, it does not have an inhibitory effect on the LT-HSC population.
ELAVL1 Knockdown Promotes Myeloid Differentiation and Inhibits In Vivo Leukemic Propagation in Human AML
Given the profound effects of ELAVL1 loss on LSCs from genetically diverse murine leukemia models, we next evaluated the functional role of ELAVL1 in human AML by introducing shScramble- (control) and shELAVL1-expressing lentiviruses into human primary AML cells (Supplementary Fig. S4A; Supplementary Tables S2 and S3). Immunophenotyping of infected cells by flow-cytometric analysis revealed increased myeloid populations, as measured by CD14+ and CD11b+ expression (Fig. 4A), as well as an overall enrichment of these antigens as demonstrated by their increased median fluorescence intensities (MFI) compared with control (Supplementary Fig. S4B). We also observed increased cell area upon ELAVL1 depletion in human primary AML compared with control cells, which is consistent with both differentiated cells that exhibit increased cytoplasmic size compared with more primitive cells as well as the differentiation phenotype observed by flow-cytometric analysis (Supplementary Fig. S4C). Furthermore, ELAVL1 reduction significantly increased cell death as measured by 7AAD+ populations (Fig. 4B). Lastly, colony-forming unit assays established postinfection revealed decreased total AML-CFU outputs in ELAVL1 knockdown conditions compared with the control (Supplementary Fig. S4D). Together, these results demonstrate that ELAVL1 plays an important role in regulating leukemic proliferation and differentiation.
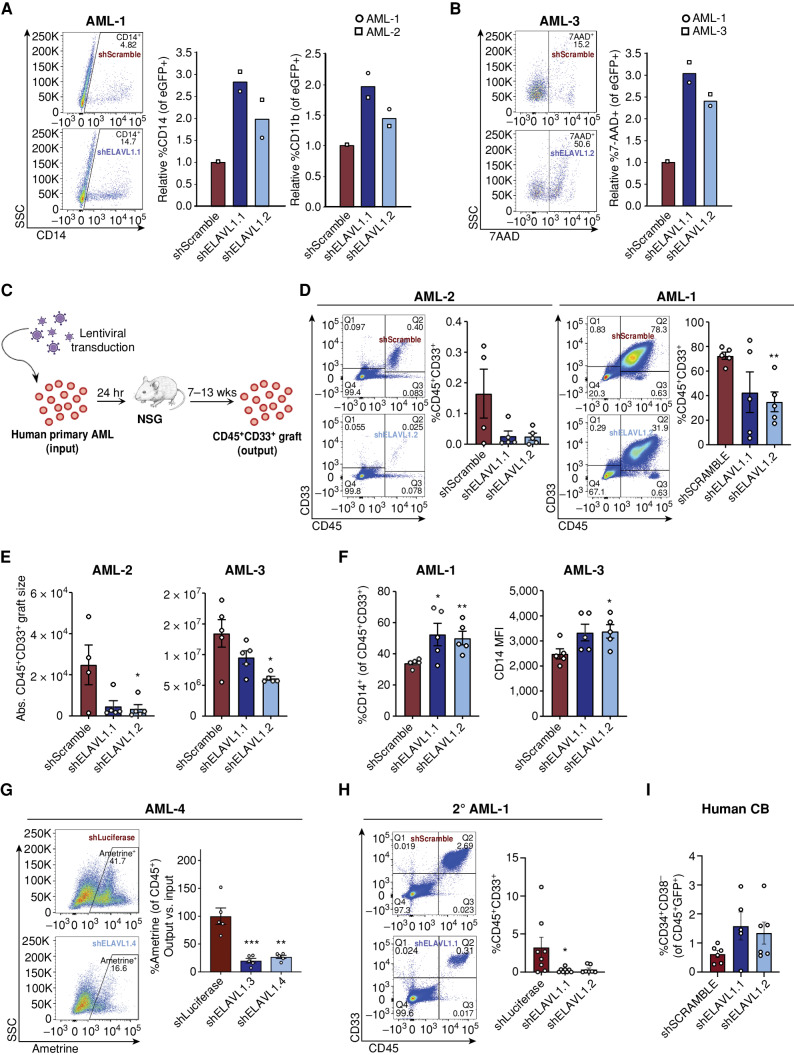
Figure 4.
ELAVL1 knockdown selectively impairs in vivo leukemic engraftment. A and B, Flow-cytometric evaluation of CD14+ and CD11b+ (A) and 7AAD+ (B) fractions of shScramble- and shELAVL1-infected primary AML cultures 10 and 2 days after infection, respectively. C, Schematic illustrating in vivo ELAVL1 loss-of-function leukemic repopulation assays. D and E, Quantitative analysis of shELAVL1-infected primary AML cells at endpoint showing %CD45+CD33+ grafts in BM (D) and absolute graft size (E) based on total cell counts in femurs and tibiae of recipient mice. F, Flow-cytometric analysis of CD14+ populations within CD45+CD33+ grafts in right femur and BM at the endpoint. G, Analysis of Ametrine+ fractions of shLuciferase- or shELAVL1-infected fractions within the injected right femur CD45+ grafts at the endpoint. H, Flow-cytometric analysis of leukemic grafts in the BM of secondary transplant recipient mice. Representative flow plots are shown. I, Percentage of human HSC in BM grafts of CB-transplanted recipient mice at the 12 weeks after transplant endpoint.*, P < 0.05; **, P < 0.01; ***, P < 0.001, determined by a two-sided Student t test. Error bars, SEM.
To directly assess the role of ELAVL1 in LSC-driven malignant propagation, we performed xenotransplantation assays using shELAVL1-infected unsorted primary AML specimens and captured the effects of ELAVL1 knockdown by measuring leukemic engraftment (CD45+CD33+) in recipient mice at endpoint (Fig. 4C). We tested three separate patient samples and in all cases knockdown of ELAVL1 significantly impaired leukemic growth in vivo as demonstrated by 50% to 80% decrease in the percentage and total number of human AML cells (Fig. 4D and E). Immunophenotyping of leukemic grafts depleted of ELAVL1 also uncovered an elevation of CD14+ (both in percentage and MFI) and CD11b+ populations (Fig. 4F; Supplementary Fig. S4E). In a fourth sample where intermediate-level infection was achieved posttransduction, the infected (Ametrine+) populations were tracked from the day of transplant (input) to endpoint (output; Fig. 4C; Supplementary Fig. S4F). At 12 weeks posttransplant, we observed a significant decrease of ∼75% to 80% in leukemic engraftment in ELAVL1-depleted AML recipient mice relative to control (Fig. 4G). To test the effects of ELAVL1 loss on LSC function, we performed the gold-standard assay of serially transplanting the BM from primary xenografts into secondary recipient mice. At the 6-week endpoint, we indeed observed a further reduction of leukemic burden in the BM of recipients of shELAVL1- relative to shScramble-transduced primary graft cells (Fig. 4H). Next, to test the effects of ELAVL1 loss on healthy HSCs, we depleted ELAVL1 in human cord blood (CB) via shRNA for in vivo hematopoietic reconstitution assays. At the endpoint, we observed that in addition to nonsignificant effects on hematopoietic engraftment (Supplementary Fig. S4G), the HSC fraction (CD34+CD38−) was not substantially altered in the BM of the recipient mice (Fig. 4I). Moreover, there was no evidence of lineage skewing as measured by expression of myeloid (CD33) and lymphoid (CD19) cell-surface markers (Supplementary Fig. S4H). Together, these data indicate that in contrast to the more modest defects of ELAVL1 loss on cultured cells from the human AML cell lines described above (Supplementary Fig. S2C–S2H), its repression in human primary AML not only substantially compromises leukemic growth in vivo but directly impairs LSC activity and thus long-term leukemic reconstitution. Moreover, the negligible effects of ELAVL1 loss on healthy long-term repopulating HSCs position this RBP as a potential therapeutic target in AML.
Inhibition of ELAVL1–RNA Interactions Selectively Impairs AML at the LSC Level
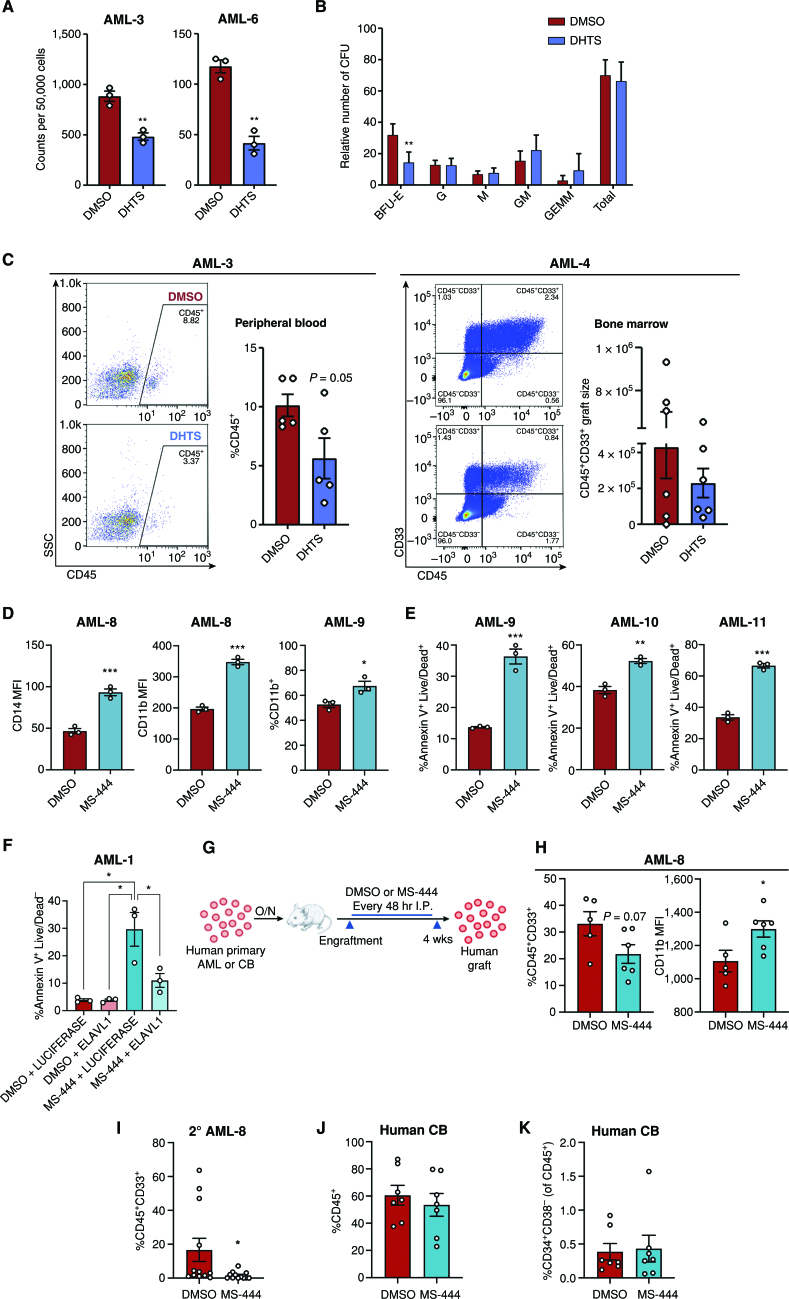
To investigate the effects of small-molecule inhibition of ELAVL1 in AML cells, we used the compound Dihydrotanshinone-I (DHTS), reported to inhibit the interaction of ELAVL1 with its mRNA targets (63). In an initial set of experiments, THP-1 cells treated with DHTS, like ELAVL1 knockout, did not significantly increase apoptosis. Moreover, they showed a trend toward myeloid maturation as indicated by elevated CD14+ populations as well as decreased cell division as determined by PKH26 labeling (Supplementary Fig. S5A–S5D). We next tested the impact of DHTS on primary AML cells in vitro. Immunophenotyping analysis of DHTS-treated primary AML cultures demonstrated significantly increased MFIs of both mature myeloid antigens, CD14 and CD11b, as well as CD33 48 hours after DHTS treatment compared with control conditions (Supplementary Fig. S5E–S5G). At the same time point, we observed a significant increase in cell death as measured by Annexin V+ 7AAD+ staining (Supplementary Fig. S5H). CFU assays carried out in the presence of DHTS yielded significantly lower malignant myeloid progenitor colonies ranging from 30% to 65% compared with vehicle control, indicating leukemic progenitors were effectively compromised (Fig. 5A; Supplementary Fig. S5I). To confirm the cell-selective context of this drug, we performed CFU assays on normal HSPC populations from lineage-depleted human umbilical CB cells. Here, the total progenitor activity remained unaltered with decreases observed only in BFU-E colonies, indicating a general insensitivity of normally committed progenitors to DHTS (Fig. 5B). Finally, to assess the functional consequences of ELAVL1 inhibition by DHTS, we treated two primary AML samples and assessed their xenotransplantation potential. Evaluating AML engraftment at 9 weeks after transplant, we observed an impairment in leukemic growth in recipient mouse peripheral blood and BM and a trend toward elevated CD14+ populations in the residual leukemic BM grafts (Fig. 5C; Supplementary Fig. S5J).
Figure 5.
Small-molecule inhibition of ELAVL1 differentially targets leukemia-propagating vs. healthy hematopoietic cells. A and B, CFU output from primary AML (A) or lineage-depleted CB cells (B) treated with DMSO or DHTS (1.1 μmol/L). n = 2 independent CB units assessed over two independent experiments, n = 3 replicates for each condition. C, Flow-cytometric analysis of leukemic grafts in peripheral blood at 9 weeks after transplant and the CD45+CD33+ graft size based on total cell counts in BM at 8 weeks after transplant. Representative flow plots are shown. D and E, Flow-cytometric analysis of myeloid maturation markers, CD14 and CD11b (D) and cell death (Annexin V+Live/Dead+; E) in human primary AML samples treated with DMSO or 5 μmol/L MS-444. F, Quantification of apoptosis within primary AML cells infected with LUCIFERASE-overexpression and ELAVL1-overexpression in the presence of DMSO or 5 μmol/L MS-444. G, Schematic illustrating in vivo administration of DMSO or MS-444 in human primary AML- or CB-engrafted mice. H, Quantitative analysis of engraftment levels (left) and CD11b expression (in the CD11b+CD45+CD33+ fraction, right) in the BM of human primary AML-engrafted mice treated with DMSO or MS-444. I, Leukemic engraftment levels of secondary recipients transplanted with BM from primary mice treated with DMSO or MS-444. J and K, Flow-cytometric analysis of hematopoietic engraftment (J) and the primitive (CD34+CD38−) HSC population (of the CD45+ graft; K) in CB-transplanted mice treated with DMSO or MS-444. *, P < 0.05; **, P < 0.01; ***, P < 0.001, determined by a two-sided Student t test. Error bars, SEM. A3SS, alternative 3’ splice site; A5SS, alternative 5’ splice site; BFU, burst-forming unit erythrocyte; G, granulocyte; GEMM, granulocyte/erythrocyte/monocyte/megakaryocyte; GM, granulocyte/monocyte; M, monocyte; MXE, mutually exclusive exons; RI, intron retention; SE, exon skipping.
To test the therapeutic potential of targeting ELAVL1, we used a bioavailable and more potent inhibitor of ELAVL1–mRNA target binding, MS-444, for a series of functional experiments. First, in vitro treatment of primary AML specimens with MS-444 significantly induced myeloid maturation as measured by increased expression of CD14+ and CD11b+ populations as well as increased cell death (Fig. 5D and E). Although other studies have validated that ELAVL1 dimerization, a process required for its ability to bind RNA targets, is directly obstructed by MS-444, which confers its specific mechanism of action in repressing ELAVL1 activity (64–66), we overexpressed ELAVL1 in the presence of MS-444 treatment to test the ability of ELAVL1 to rescue the cellular effects of MS-444 as a further validation of inhibition specificity in AML cells. Indeed, we observed a significant rescue of MS-444's apoptotic phenotype by ELAVL1 overexpression (Fig. 5F; Supplementary Fig. S5K), confirming that MS-444 functionally inhibits ELAVL1 in this context. Next, we performed in vivo leukemic reconstitution assays with the administration of MS-444 to test the effects of ELAVL1 inhibition on leukemic propagation. Upon engraftment, mice were treated with 20 mg/kg of MS-444 or vehicle control intraperitoneally (i.p.) every 48 hours for 4 weeks (Fig. 5G). At endpoint, the BM from the MS-444–treated recipients showed decreased leukemic burden and significantly increased myeloid differentiation as measured by CD11b+ expression compared with vehicle control recipients (Fig. 5H). To assess the effects of MS-444–driven inhibition of ELAVL1 on LSCs, we serially transplanted the BM from primary mice into secondary recipients in two cell doses. At the endpoint, the mice that received the MS-444–treated BM demonstrated a dramatic impairment of leukemic reconstitution in which half of the mice did not have a leukemic graft at all (Fig. 5I). In addition, using limiting dilution analysis to evaluate LSC frequency differences by virtue of binary engraftment calls in low versus high cell dose-transplanted recipients, we determined that LSCs decreased in MS-444–treated mice by 83% in comparison with vehicle-treated controls (Supplementary Table S4). Together, this demonstrates that ELAVL1 inhibition by MS-444 significantly impairs the ability of LSCs to drive long-term leukemic propagation.
Finally, to evaluate the effects of MS-444 on normal tissues and primitive normal hematopoietic cells in particular, we treated human CB-engrafted mice with the same in vivo regimen of MS-444 (Fig. 5G). Here we observed no evidence of toxicity with all animals being healthy and showing no evidence of adverse reactions throughout the entirety of the treatment regimen, as was observed in our MS-444–treated leukemic xenografts. Importantly, the CB grafts of MS-444–treated recipients showed no change in their levels compared with vehicle-treated mice and the HSPC (CD34+) and most primitive HSC-enriched populations (CD34+CD38−) were also preserved at control levels in the grafts of MS-444–treated recipients (Fig. 5J and K; Supplementary Fig. S5L). Moreover, MS-444–treated grafts were normal in all respects and exhibited no evidence of lineage skewing (Supplementary Fig. S5M). Together, these data demonstrate not only the therapeutic potential of small-molecule inhibition of ELAVL1 to treat AML but also the selectivity of this strategy in targeting the LSC population known to drive long-term propagation of the AML.
ELAVL1 Enacts LSC-Supportive Posttranscriptional Circuitry
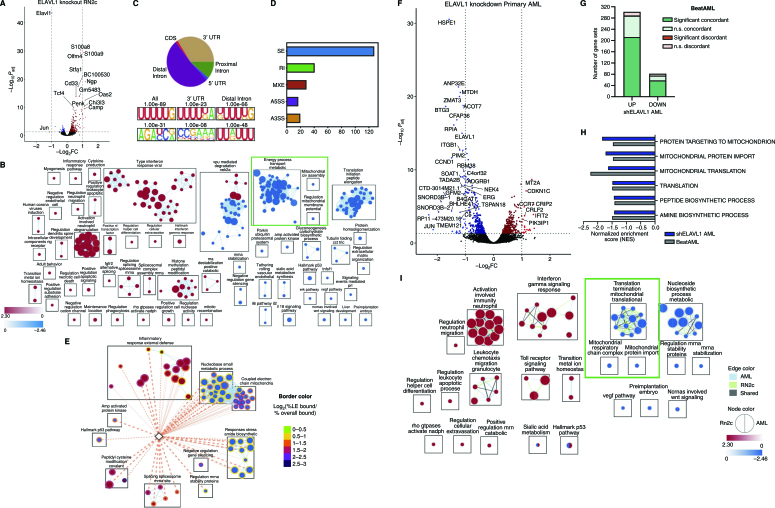
Given our findings that ELAVL1 is essential for LSC maintenance and its known role as a stabilizer of its RNA targets, we sought to comprehensively examine ELAVL1's underlying mechanism using global transcriptomic profiling upon its knockout in the LSC-rich RN2c cells. ELAVL1 knockout resulted in 243 upregulated and 47 downregulated transcripts (Fig. 6A; Supplementary Table S5). To capture the full spectrum of coordinated changes in functionally related processes influenced by ELAVL1 loss, we performed GSEA. Consistent with the functional impairment of LSCs in our ELAVL1-depleted primary AML xenografts, we uncovered 252 enriched gene sets and identified that, in particular, signatures of LSC maintenance are diminished whereas those of myeloid differentiation are activated with ELAVL1 loss (Supplementary Fig. S6A). Enrichment mapping of ELAVL1-dependent transcriptional changes furthermore revealed that the most significant negatively regulated clusters relate to metabolism, cytoplasmic translation, and mitochondrial respiration (Fig. 6B; Supplementary Table S6). The outcome of our transcriptome profiling thus highlights the possibility that the role of ELAVL1 in LSC maintenance involves reprogramming of certain cellular metabolic processes.
Figure 6.
Characterization of the ELAVL1-dependent circuitry in primitive leukemic cells. A, Volcano plot of differential gene expression in ELAVL1-knockout RN2c RNA sequencing (RNA-seq) Genes with significant differences in expression are highlighted. Blue and red dots represent genes significantly downregulated or upregulated, respectively, using a Padj < 0.05 (RNA-seq) cutoff. B, Enrichment map of gene sets significantly enriched (FDR < 0.1) in the transcriptome of ELAVL1-knockout RN2c cells. C, Distribution of ELAVL1 eCLIP peaks in different genic regions (top) and most common ELAVL1-binding motif sequences (bottom) in mouse bcCML cells. D, Distribution of splicing events in ELAVL1-knockout RN2c cells. E, Enrichment map of pathways enriched (FDR < 0.1) in the ELAVL1-knockout RN2c transcriptome and containing >5% of leading-edge transcripts bound by ELAVL1. Color of borders is based on the enrichment of transcript binding to leading edge relative to gene set background. F, Volcano plot of differential gene expression in ELAVL1-KD human primary AML. Blue and red dots represent genes significantly downregulated or upregulated, respectively, using a Padj < 0.05 cutoff. G, Number of pathways in the human ELAVL1-knockdown AML transcriptome that are significantly or nonsignificantly concordant and discordant in the BeatAML RNA-seq data set. H, Normalized enrichment scores (NES) of downregulated mitochondrial gene sets in the human ELAVL1-knockdown RNA-seq data set (highlighted by the green box in Supplementary Fig. S6I). I, Enrichment map of gene sets significantly (FDR < 0.25) altered in both ELAVL1-knockdown human AML and ELAVL1-knockout RN2c transcriptomes.
We next sought to identify direct LSC-specific binding targets of ELAVL1 by performing enhanced cross-linking immunoprecipitation followed by deep-sequencing (eCLIP-seq; ref. 67). To enable efficient eCLIP pulldown, we used mouse bcCML cells, which similarly to RN2c are enriched in ELAVL1-dependent LSCs (Supplementary Fig. S3B–S3E), while importantly exhibiting the requisite high expression of ELAVL1 protein (Supplementary Fig. S6B). We discovered 4,345 significant ELAVL1-binding peaks across 1,548 genes with a preference for intronic regions, suggesting that ELAVL1 can act in the nucleus on pre-mRNA species, as well as 3′UTRs, consistent with typical binding profiles underlying its role in mRNA stabilization (Fig. 6C; Supplementary Table S7). In agreement with this, we identified nuclear and cytoplasmic localization of ELAVL1 in both mouse leukemic BM and human primary AML cells (Supplementary Fig. S6C). In line with previous findings (68, 69), an enrichment for U-rich binding motifs was identified within regions bound by ELAVL1 (Fig. 6C). Lastly, biological processes overrepresented among ELAVL1-bound transcripts included hematopoietic differentiation, transcriptional control, mRNA processing, and mRNA splice regulation (Supplementary Table S7).
To identify transcripts directly regulated by ELAVL1, we interrogated the expression outcomes of ELAVL1-bound transcripts from the eCLIP-seq in the ELAVL1-depleted transcriptome. From this, we identified 152 genes whose significant differential transcript levels appear directly linked to physical association with ELAVL1 (Supplementary Fig. S6D; Supplementary Table S7). Upon this integrative analysis, we tested by qRT-PCR whether MS-444 induces similar expression changes of ELAVL1's downstream targets, MYC and NRF1, both bound and significantly downregulated by ELAVL1 loss, and IRF7, a target significantly upregulated in our RNA sequencing (RNA-seq) data set. Indeed, we observed significant dysregulation of these targets consistent with the changes observed in our RNA-seq, highlighting that MS-444 imparts key transcriptomic changes similar to ELAVL1 loss (Supplementary Fig. S6E). Furthermore, the 152 genes bound by ELAVL1 and whose expression levels were dependent upon it were predominantly upregulated in response to ELAVL1 loss (Supplementary Fig. S6D), a significant trend found amongst all bound transcripts (Supplementary Fig. S6F). Within the bound and downregulated transcripts upon ELAVL1 loss were several LSC enforcers and oncogenes including Gpr56, Dazap1 and the previously appreciated direct target of ELAVL1, Myc (refs. 70–75; Supplementary Fig. S6D). In contrast, the direct ELAVL1 targets upregulated upon its loss include Neat1, a differentiation promoter and a known ELAVL1 target (76, 77), as well as a significant overrepresentation of mRNA splicing regulators (Supplementary Fig. S6D and S6G; Supplementary Table S7). Noting this latter class of targets as well as the pre-mRNA binding action of ELAVL1 (Fig. 6C; Supplementary Fig. S6G), we explored mRNA splicing changes upon ELAVL1 depletion. This analysis revealed 230 diverse changes with exon skipping being the most common event (Fig. 6D; Supplementary Table S8). Interestingly, among these exon-skipping events, we again identified dysregulated mitochondrial genes. More specifically, we noted the presence of several altered exon-splicing events [delta percent spliced in (PSI) > 0.25] in genes with characterized roles in mitochondrial function and integrity including Ewsr1, Samd8, Nsun3, Pam16, and Mrtfl1 (Supplementary Fig. S6H).
Given that our multiomics analyses implicate mitochondrial regulation as a probable hub downstream of ELAVL1, we examined whether ELAVL1 binding contributes to the direct regulation of mitochondrial genes. Overall, we found that ELAVL1 is bound to leading-edge transcripts in 85% of the significantly enriched gene sets observed in the ELAVL1 knockout transcriptome, including mRNA splicing, activated immune functions, and metabolic processes (Fig. 6E). We noted that an average of 9.3% (range, 5.7%–15%) of the leading-edge transcripts in 14 gene sets related to the electron transport chain are also bound by ELAVL1, a significant 1.6-fold enrichment over the gene set background (t test P < 0.05; Fig. 6E).
Altogether, profiling of the ELAVL1-directed regulon via integrated analyses of the RNA-interactome and transcriptome strongly implicates mitochondrial activity as a critical axis through which ELAVL1 supports LSCs.
ELAVL1 Repression Impairs Leukemic Mitochondrial Function
We next aimed to profile ELAVL1 dependencies integral to the maintenance of human AML. To this end, we performed RNA-seq in primary patient AML cells upon ELAVL1 knockdown and identified 333 upregulated and 376 downregulated genes (Fig. 6F; Supplementary Table S9). To assess the clinical relevance of these ELAVL1-associated programs, we compared the shELAVL1 transcriptome to RNA-seq data of 494 bulk AML samples from the BeatAML clinical data set (78). We observed a positive correlation in overall gene-expression profiles of below-median ELAVL1 expressers and ELAVL1-knocked down AML, particularly among differentially expressed genes (ρ = 0.32, P < 10e−10). Assessing pathway-level changes in the shELAVL1 transcriptome, we again observed dysregulation of LSC maintenance and myeloid differentiation gene signatures (Supplementary Fig. S6I; Supplementary Table S9), demonstrating ELAVL1's role in maintaining stemness features in human AML. Overall, we discovered 410 aberrantly regulated pathways, of which 70% are significantly concordant with the BeatAML data set with no discordant events (Fig. 6G; Supplementary Fig. S6J). Most importantly, echoing our findings in the LSC-rich RN2c setting, even when profiled at the bulk level, we observed a negative enrichment of mitochondrial import and translation in AML samples with experimentally or disease-specific reduced levels of ELAVL1 (Fig. 6H; Supplementary Fig. S6J). The strong mirroring of these and other expression signatures suggests that ELAVL1-dependent programming actively shapes the transcriptomic landscape of clinical AML.
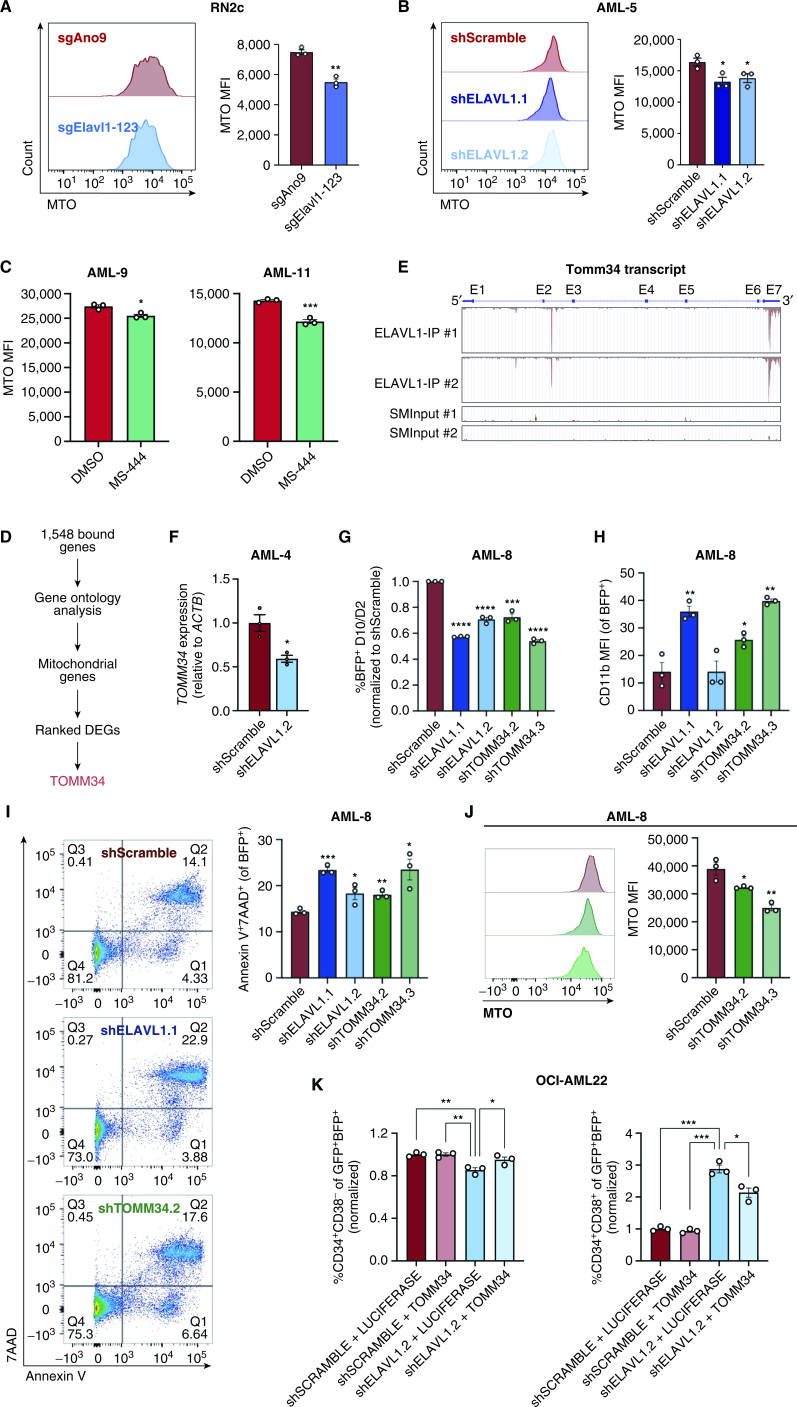
Finally, in an effort to uncover core ELAVL1-mediated biological processes underlying its support of human LSC, we compared pathway-level changes in ELAVL1-depleted bulk human AML and murine LSC-rich RN2c cells. Within commonly altered gene sets, we again confirmed the recurring theme of activation of myeloid differentiation programs, and repression of constituents of mitochondrial function and integrity (Fig. 6I). Given that dependence on mitochondrial function has emerged as a selective regulatory mechanism of the LSC compartment (79), we hypothesized that ELAVL1's essential role in LSCs may be mediated by its control over mitochondrial processes. Indeed, flow-cytometric measurements of mitochondrial activity via MitoTracker dye staining showed significant decreases in both murine RN2c and human primary AML cells upon ELAVL1 depletion (Fig. 7A and B; Supplementary Fig. S7A and S7B). Similarly, treatment of human primary AML specimens with MS-444 significantly impaired mitochondrial function as compared with the vehicle control (Fig. 7C). These results, in combination with our comprehensive molecular profiling support that maintenance of mitochondrial activity is an important mechanism through which ELAVL1 achieves its critical and selective role in supporting AML LSCs.
Figure 7.
TOMM34 is a direct effector of ELAVL1 and is essential for mitochondrial metabolism and maintenance of primitive AML cells. A–C, Quantification of MitoTracker Orange (MTO) MFI in ELAVL1-depleted RN2c (A) and human primary AML cells (B) and in human primary AML cells treated with DMSO or 5 μmol/L MS-444 (C) 72 hours after infection or treatment. n = 3 technical replicates for each experiment. D, Flow chart illustrating the steps in identifying a top downregulated mitochondrial gene directly bound and regulated by ELAVL1. E, UCSC Genome Browser tracks showing ELAVL1-binding peaks along the TOMM34 transcript in reference to size-matched small input (SMInput) controls. F, qPCR of TOMM34 in shScramble- and shELAVL1-infected human primary AML. G–I, Flow-cytometric analysis of proliferation (BFP+; G), myeloid differentiation (H), and cell death (I) in ELAVL1- and TOMM34-depleted human primary AML cells compared with controls. J, MTO analysis of TOMM34-depleted human primary AML. K, Flow-cytometric evaluation of LSC (CD34+CD38−, left) and committed progenitor cells (CD34+CD38+, right) within OCI-AML22 cells coexpressing shSCRAMBLE/shELAVL1 and LUCIFERASE-/TOMM34-overexpression. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, as determined by a two-sided Student t test. Error bars, SEM.
TOMM34 Is a Direct Effector of ELAVL1 in AML
Given that our integrative bioinformatic analyses and functional experiments demonstrate that mitochondrial control is a key axis through which ELAVL1 drives its phenotype in AML, we sought to identify an ELAVL1-direct effector that might underlie this control. To do this, we returned to the list of 1,548 transcripts bound by ELAVL1 as identified by eCLIP-seq and using Gene Ontology analysis isolated mitochondrial genes and ranked them based on their significance of differential expression in the human AML RNA-seq data set (Fig. 7D). This analysis identified Translocase of Outer Mitochondrial Membrane 34 (TOMM34) as the ELAVL1-directly bound mitochondrial gene with the topmost downregulation upon ELAVL1 loss (Fig. 7D and E). Indeed, upon depletion of ELAVL1 in human primary AML, TOMM34 is significantly downregulated as measured by qRT-PCR (Fig. 7F). TOMM34 is a cochaperone that facilitates the heat shock protein HSP70/HSP90-mediated import of mitochondrial preproteins (80–82), and although it has been associated with poor survival in solid tumors (83, 84), its physiologic role in mitochondrial metabolism and involvement in leukemia pathogenesis is unknown. Interestingly, however, TOMM34 expression follows a similar pattern to ELAVL1, with its transcripts significantly enriched in the LSC+ fraction of human AML (28), increasing expression across progressively aggressive phases of CML (46), preferential expression at the protein level in the LSC compartment of the patient-derived OCI-AML-8227 AML model (47), and more significant expression in relapsed AML samples compared with their paired diagnosis counterparts (refs. 3, 55, 56; Supplementary Fig. S7C–S7E). Opposingly, TOMM34 is expressed at lower levels in HSCs from human BM compared with downstream progenitor cells (Supplementary Fig. S7F), much like the expression profile observed for ELAVL1 (Fig. 2I). Moreover, TOMM34 transcript levels also correlate with ELAVL1 expression in the diverse BeatAML data set of primary AML samples, consistent with the predicted role of ELAVL1 in stabilizing TOMM34 by virtue of ELAVL1's association with its 3′UTR (Supplementary Fig. S7G).
To determine the functional similarity of TOMM34 to ELAVL1, we performed immunophenotyping and apoptosis assays upon shRNA-mediated TOMM34 depletion in human primary AML in parallel to ELAVL1 knockdown. Here, we observed that repression of TOMM34 significantly impairs proliferation and promotes myeloid maturation and cell death, mirroring closely the phenotype observed by ELAVL1 loss (Fig. 7G–I; Supplementary Fig. S7H and S7I). To assess the effects of TOMM34 loss on mitochondrial function, we used MitoTracker Orange in TOMM34-depleted human primary AML cells and show that, as we observed upon ELAVL1 loss, mitochondrial activity is indeed significantly compromised compared with controls (Fig. 7J). Finally, to determine whether TOMM34 is a direct effector of ELAVL1, we tested the ability of TOMM34 overexpression to rescue cellular phenotypes induced by ELAVL1 loss, specifically in the LSC compartment. To do this, we simultaneously overexpressed TOMM34 and depleted ELAVL1 via shRNA in the primary AML-derived cell line OCI-AML-22, which maintains a functional hierarchy of LSC+ populations (CD34+CD38−) and downstream progenitor cells (CD34+CD38+). Flow-cytometric analysis of double-infected populations showed significant rescue of reduced LSC+ fractions, as well as rescue of early progenitor commitment as measured by CD34+CD38+ cells (Fig. 7K; Supplementary Fig. S7I and S7J). Altogether, these data indicate that TOMM34 is a positively regulated direct downstream target of ELAVL1 through which it enacts its role in maintaining mitochondrial function for AML survival.
DISCUSSION
Elucidation of LSC-targeted therapies remains a profound unmet need for AML, a cancer that under standard-of-care therapy is characterized by high rates of relapse and poor long-term survival. Current efforts to uncover key molecular LSC dependencies have largely overlooked the potentially target-rich class of posttranscriptional regulators. In vivo functional genetic screens have been important in showcasing the insights that come from interrogating cancer cell dependencies in the setting of a complex in situ niche, however, have yet to be applied in such a way as to uncover cancer stem cell–specific regulators. We describe the first LSC-focused pooled CRISPR dropout screen in AML performed uniquely in the in vivo serial transplantation setting. This approach has uncovered regulators of clear importance to LSCs and thus may serve as a novel strategy the field can capitalize on to prioritize candidates of maximum clinical interest more systematically. Moreover, with a focus on posttranscriptional regulators as an understudied class of candidate LSC determinants, this unique approach identified RBPs essential to the repopulating and/or self-renewal function of LSCs. Of the 128 RBPs preferentially expressed in LSCs that we systematically screened, 32 were required for in vivo leukemic propagation. This identification of a large number of RBPs underlying LSC function combined with the enrichment in expression of the entire class of RBPs in LSCs speaks to an intriguing dependence of LSCs on RBP-driven posttranscriptional control mechanisms that warrant future in-depth investigations into their therapeutic potential. This list of RBPs is diverse and encompasses regulators known to influence virtually all aspects of RNA metabolism. In addition, our screen has highlighted specific cellular pathways known to be under the control of certain hit RBPs, which have been previously implicated in LSCs (e.g., rRNA metabolism; ref. 85), and others including tRNA modification and ribosome biogenesis, not before appreciated for their specific contribution to AML LSC function. Given the parallels between LSCs and other tissue-specific cancer stem cells (2), our findings portend value in exploring the extent to which RBP-mediated regulation contributes to the functionality of cells driving diverse cancer types.
Of the identified RBP hits in our screen, ELAVL1 imparted amongst the most significant inhibitory effects to LSCs and leukemic cell growth over the course of the screen and when independently targeted was essential for mouse and human leukemic reconstitution. Overexpression of ELAVL1 has been reported in various solid tumor types (86–88), as well as myeloid and lymphoid leukemias (46, 89, 90). However, its role in the stem cell compartment of leukemia has thus far not been addressed. Our functional evaluation in combination with transcriptional profiling following ELAVL1 knockout in an LSC-rich context indicates that ELAVL1 enforces a larger molecular profile that maintains LSC stemness, restricts differentiation and preserves survival. Importantly, we find that ELAVL1 disruption via shRNA or small-molecule interventions significantly impairs LSC self-renewal but allow for a relative sparing of the compartment of normal mouse and human HSPCs, raising the significance of this RBP as a candidate therapeutic target in AML. Our results with DHTS and MS-444 further suggest that inhibition of RBP–mRNA interactions might also be as effective as depletion of the RBP itself, a concept also hinted at by the Pfam targeting strategy of our screen. As many noncanonical RBPs have functions beyond regulation of RNA (91, 92), explicit reliance on their RNA-binding features for LSC function may also provide novel opportunities for antileukemic intervention. Indeed, this concept is exemplified by our results with MS-444 administered in vivo, which demonstrate the clear therapeutic potential and relevance of targeting ELAVL1–mRNA interactions to directly impair LSC function and reduce leukemic burden.
ELAVL1 has well-described roles in stabilizing pro-cell growth mRNAs through association with AU-rich regions within 3′UTRs (69, 93–96). Although ELAVL1 has also been described to influence splicing in nonhematopoietic tissues (97–99), as RBPs generally confer distinct cell-context–specific effects on the global splicing status, the nature of any splicing regulation by ELAVL1 in AML LSCs has not been known (100). In LSCs, we find that the majority of ELAVL1's binding events are intronic, in line with the findings of Mukherjee and colleagues, who documented a previously unrecognized importance for intronic ELAVL1 binding in pre-mRNA stabilization (69). In addition, we reveal that 15 ELAVL1-bound transcripts appear dependent on ELAVL1 for stabilization as compared with 141 whose expression is elevated upon ELAVL1 deletion. Among the latter ELAVL1-bound and negatively regulated transcripts, we also observed an enrichment in regulators of splicing. Together, these findings not only indicate that a much larger pool of total bound transcripts exists for which ELAVL1 interaction may be repressive in these cells, but that this noncanonical role of ELAVL1 is selective to splicing regulators. Consistent with this, we found that global splicing was indeed significantly altered upon ELAVL1 knockout, which supports a critical role for ELAVL1 in propagating a specific LSC alternative splicing (AS) program both directly and indirectly. These findings are intriguing given the burgeoning appreciation for AS dysregulation as a result of splicing regulator mutations that promote leukemic transformation, propagation, and relapse (101–104) and the potential for altered expression of splicing regulators to promote pathogenic splicing in other cancers (105–107).
Our integrative omics analysis uncovered an ELAVL1-nucleated posttranscriptional circuitry in LSCs that in large part coalesces on a signature of oxidative phosphorylation conservation. In addition to being a previously uncharacterized target of ELAVL1, this finding is particularly interesting in light of studies that situate mitochondrial metabolism as a critical axis that LSCs are selectively dependent on relative to normal HSCs. More specifically, AML and CML LSCs maintain a decreased spare reserve glycolytic capacity in comparison with HSCs rendering them especially vulnerable to inhibitory strategies that target mitochondrial metabolism. Such strategies, including targeting of mitochondrial protein synthesis, mitochondrial DNA replication, amino acid metabolism, or mitochondrial protein degradation, selectively kill LSCs while sparing HSCs (6, 108, 109). Intriguingly, even among the most alternatively spliced transcripts in ELAVL1-depleted LSCs were regulators of mitochondrial function, providing the first link between not only RNA splicing, but a specific ELAVL1-mediated AS program and mitochondrial metabolism in LSCs. Furthermore, we identify for the first time TOMM34 as not only a key effector of ELAVL1 in human primary AML, but offering the possibility of mitochondrial import as a novel avenue through which LSC metabolism may be therapeutically targeted. TOMM34 is a cochaperone that functions as the gateway mediating mitochondrial protein import through the translocase of outer mitochondrial (TOM) complex by stabilizing nuclear-encoded, mitochondrially destined pre-proteins in an unfolded state. Once shuttled across the TOM complex, these highly diverse preproteins, which encode the majority of proteins necessary for mitochondrial function, then undergo more selective sorting and localization to specific membranes and regions within the mitochondria (110). As such, targeting of mitochondrial import at the level of the ELAVL1–TOMM34 axis may serve as a more pervasive approach to disrupting localization and ultimately function, of a greater variety of mitochondrial proteins and thus offer the potential to solidify a robust inhibition of mitochondrial processes necessary for LSC maintenance and AML survival. Considered together, our findings here highlight a unique dependence of genotypically distinct primitive leukemic cells on oxidative metabolism that our data indicate can be counteracted by interfering with its posttranscriptional control via ELAVL1. Our work further provides insight into a clinically relevant connection between RBP-mediated post-transcriptional regulation and mitochondrial metabolism in AML LSCs that to our knowledge has not been elucidated to date.
Altogether, our combined functional and molecular analyses showcase ELAVL1 as a critical novel regulator of LSCs that utilizes a combination of changes to the RNA landscape to coordinately enforce a state that supports LSC-optimal mitochondrial metabolism. Together with other diverse RBP regulators identified in our LSC-directed in vivo CRISPR screen, these findings highlight stem cell–adapted in vivo screening as a tractable tool to identify high-value therapeutic targets, establish RBPs as essential players in LSC biology, and open the door to elucidating and therapeutically exploiting their mechanisms of action.
METHODS
Mouse Maintenance and Transplants
B6.SJL (Ly5.1+, RRID: IMSR_JAX:002014), C57Blk/6 (Ly5.2+, RRID: IMSR_JAX:000664), and NSG (RRID: IMSR_JAX:005557) mice were bred and maintained at McMaster University and the University Health Network. All animal experiments were performed in accordance with institutional guidelines approved by the institutional Animal Research Ethics Boards. Twenty-four hours prior to transplantation by tail vein or intrafemoral injection, mice were sublethally irradiated (1 × 580 Rad or 1 × 315 Rad). BM and spleen were harvested from moribund mice, crushed in RPMI + 10% FBS, and passed through 40-μm cell strainers (Corning; cat. no. 352340). Ammonium Chloride (STEMCELL Technologies; cat. no. 07850) was used for lysis of red blood cells.
Leukemia and Immortalized Cell Lines
RN2c cells (MLL-AF9/NrasG12D/hCas9; a kind gift from Dr. Vakoc, Cold Spring Harbor Laboratory; received in 2015) were cultured in RPMI supplemented with 10% FBS, at a maximum density of 1 million cells per mL. MLL-AF9 and bcCML cell lines were generated as described (30, 58) and cultured in SFEM (STEMCELL Technologies; cat. no. 09650) supplemented with 20 ng/μL mouse SCF (R&D Systems; cat. no. 455-MC-010), 10 ng/μL mouse IL3 (R&D Systems; cat. no. 403-ML-050) and 10 ng/μL mouse IL6 (R&D Systems; cat. no. 406-ML-025). RN2c, MLL-AF9, and bcCML cells were all used at approximate passage number in vivo of 2–3. 293FT (RRID: CVCL_6911, acquired from ThermoFisher in 2012) and HeLa (RRID: CVCL_0030, acquired from ATCC in 2010) were cultured in DMEM supplemented with 10% FBS. THP-1 (RRID: CVCL_0006, obtained from ATCC in 2015) cells were cultured in RPMI supplemented with 10% FBS. 293FT, HeLa, and THP-1 cells were passaged for 2 to 4 passages before use. Cell line authentication and mycoplasma testing for all lines were last performed in 2020.
Culture of Primary AML Patient Samples
All AML patient samples (n = 11) were obtained as peripheral blood draws with written informed consent and conducted in accordance with recognized ethical guidelines by the Research Ethics Boards at McMaster University (HiREB # 08-042T) and University Health Network (UHN) Research Ethics Board (CAPCR # 20-6026) in accordance with Canadian Tri-Council Policy Statement on the Ethical Conduct for Research Involving Humans (TCPS). Immediately following harvest samples were subjected to Ficoll-Paque PREMIUM (Cytiva; cat. no. 17544203) separation, mononuclear cells were stored in the vapor phase of liquid nitrogen in 10% DMSO, 40% FCS and alpha MEM. Primary samples were thawed in X-VIVO (Lonza; cat. no. BEBP04-743Q) 50% FBS with 100 μg/mL DNAse prior to using in in vitro and in vivo assays. Primary AML samples were grown in AML growth media consisting of X-VIVO with 20% BIT Serum Substitute (STEMCELL Technologies; cat. no. 09500) or StemSpan SFEM II (STEMCELL Technologies; cat. no. 09655), supplemented with 100 ng/mL human SCF (R&D Systems; cat. no. 255-SC-050), 10 ng/mL human IL3 (R&D Systems; cat. no. 203-IL-050), 20 ng/mL human IL6 (PeproTech; cat. no. AF-200-06), 20 ng/mL human TPO (PeproTech; cat. no. AF-300-18), and 100 ng/mL human FLT3 L (R&D Systems; cat. no. 308-FKN-100).
Lentiviral-Infected and DHTS-Treated Primary AML Transplantation Assays
Production of shELAVL1 and shLuciferase/shScramble expressing lentiviral particles was performed as previously described (111) and briefly described under “Lentivirus Production” below and validated by qRT-PCR and/or western blot. For knockdown experiments, AML cells were thawed and transduced at an MOI of 50 for 24 or 48 hours, depending on the sample. For drug treatment experiments, AML cells were thawed and cultured with 1.1 μmol/L DHTS (Millipore Sigma; cat. no. D0947-10MG) or equivalent volume of vehicle DMSO (Fisher Scientific; cat. no. BP231-100). All cells were transplanted intrafemorally into sublethally irradiated (315 Rad) NSG mice (RRID: IMSR_JAX:005557) at their corresponding time points. Mice were sacrificed 9 to 14 weeks after transplant, and BM from the right femur (site of injection) and remaining tibias, pelvis, and left femur were harvested along with spleens, filtered, and red blood cell lysed using ammonium chloride (STEMCELL Technologies; cat. no. 07850). Reconstituted mouse BM and human AML was blocked with mouse BD Fc Block (BD Biosciences; cat. no. 553142, RRID: AB_394657) and human IgG (MilliporeSigma; cat. no. I4506, RRID: AB_1163606), respectively. Cells were subsequently stained with fluorochrome-conjugated antibodies: CD45 Pacific Blue (Thermo Fisher Scientific; cat. no. MHCD4528, RRID: AB_10375161) or BV421 (BD Biosciences; cat. no. 563879, RRID: AB_2744402); CD33 PE (BD Biosciences; cat. no. 347787, RRID: AB_400350); CD14 PE-Cy7 (BD Biosciences; cat. no. 561385, RRID: AB_10611732) or APC-H7 (BD Biosciences; cat. no. 561384, RRID: AB_10611720); CD11b BV605 (BD Biosciences; cat. no. 562721, RRID: AB_2737745); and 7AAD PerCP-Cy5.5 (BD Biosciences; cat. no. 559925, RRID: AB_2869266) for quantitative analysis by flow cytometry.
Primary AML Immunophenotyping and Apoptosis
For knockdown experiments, AML cells were thawed, transduced at an MOI of 50 with lentivirus expressing pLKO.1-BFP-shScramble, -shELAVL1.1 (5′-GCAGCATTGGTGAAGTTGAAT-3′), or -shELAVL1.2 (5′-CCCATCACAGTGAAGTTTGCA-3′) and cultured for 10 days. For DHTS experiments, AML cells were thawed and cultured with 5.4 μmol/L DHTS (MilliporeSigma; cat. no. D0947-10MG) or equivalent DMSO (Fisher Scientific; cat. no. BP231-100) volume for 48 hours. At their corresponding time points, cells were blocked with human IgG (MilliporeSigma; cat. no. I4506, RRID: AB_1163606) and subsequently stained for quantitative flow-cytometric analysis. For evaluation of knockdown experiments: CD14 PE (BD Biosciences; cat. no. 347497, RRID: AB_400312); CD11b APC (BD Biosciences; cat. no. 550019, RRID: AB_398456); and 7AAD PerCP-Cy5.5 (BD Biosciences; cat. no. 559925, RRID: AB_2869266). For DHTS experiments: CD33 PE (BD Biosciences; cat. no. 347787, RRID: AB_400350) or BV605 (BD Biosciences; cat. no. 745229, RRID: AB_2742818); CD14 FITC (BD Biosciences; cat. no. 347493, RRID: AB_400311) or APC-H7 (BD Biosciences; cat. no. 561384, RRID: AB_10611720); CD11b BV605 (BD Biosciences; cat. no. 562721, RRID: AB_2737745); and Annexin V AlexaFluor-647 (Innovative Research; cat. no. A23204, RRID: AB_2341149). For all in vitro primary AML treated with MS-444 experiments, AML cells were thawed and cultured with 5 μmol/L MS-444 (MedChemExpress; cat. no. HY-100685) or equivalent DMSO (Fisher Scientific; cat. no. BP231-100) volume for 4 or 7 days. At their corresponding time points, cells were blocked with human IgG (MilliporeSigma; cat. no. I4506, RRID: AB_1163606) and subsequently stained for quantitative flow-cytometric analysis: Annexin V PE-CF594 (BD Biosciences; cat. no. 563544, RRID: AB_2869502) or AlexaFluor-647 (Innovative Research; cat. no. A23204, RRID: AB_2341149); Live/Dead Fixable stain Yellow (Thermo Fisher Scientific; cat. no. L34959), Blue (Thermo Fisher Scientific; cat. no. L23105), or Green (Thermo Fisher Scientific; cat. no. L23101); CD14 FITC (BD Biosciences; cat. no. 347493, RRID: AB_400311) or APC-H7 (BD Biosciences; cat. no. 561384, RRID: AB_10611720); and CD11b BV605 (BD Biosciences; cat. no. 562721, RRID: AB_2737745) or BUV395 (BD Biosciences cat. no. 563839, RRID: AB_2716869).
Isolation of Human Cord Blood Hematopoietic HSPCs
All human umbilical CB samples (n = 4) were obtained with written informed consent and conducted in accordance with recognized ethical guidelines by the Research Ethics Boards at McMaster University (HiREB # 14-527-T) and UHN (REB # 20-6026) in accordance with Canadian Tri-Council Policy Statement on the Ethical Conduct for Research Involving Humans (TCPS). Freshly harvested CB samples were stored for a maximum of 1 day after collection at 4°C and then mononuclear cells were collected by centrifugation with Ficoll-Paque PREMIUM (Cytiva; cat. no. 17544203), followed by red blood cell lysis with Ammonium Chloride (STEMCELL Technologies; cat. no. 07850). Cells were subsequently stained with a cocktail of lineage-specific antibodies (CD2, CD3, CD11b, CD11c, CD14, CD16, CD19, CD24, CD56, CD61, CD66b, and GlyA; STEMCELL Technologies; cat. no. 19356) for negative selection of lineage-depleted (Lin−) cells using an EasySep immunomagnetic column (STEMCELL Technologies; cat. no. 18000). Live cells were discriminated on the basis of cell size, granularity and, as needed, absence of viability dye 7-AAD (BD Biosciences; cat. no. 559925, RRID: AB_2869266) uptake and stored as Lin− cells in the vapor phase of liquid nitrogen in 10% DMSO + 90% FBS.
AML and CB Clonogenic Progenitor Assays
Thawed primary AML samples were counted and plated in a methylcellulose-based hematopoietic colony formation medium (Colony Gel, ReachBio cat. no. 1102), supplemented with 1.1 μmol/L DHTS (MilliporeSigma; cat. no. D0947-10MG) or equivalent DMSO (Fisher Scientific; cat. no. BP231-100) volume. Colonies were scored on days 10 to 14. Human CB samples were plated as described above with a density of 1 × 103 cells per 35 mm plate. Cell suspensions were plated in duplicate, and loose colonies consisting of 10 or more cells were scored and counted. THP-1 (RRID: CVCL_0006) cells were plated at a density of 200,000 and 1,000 cells/mL, respectively. Putative LSC-enriched populations were isolated from freshly expanded MLL-AF9 mouse leukemia cells by sorting c-Kithigh (top 10%) cells on a MoFlo XDP cell sorter (Beckman Coulter). Sorted cells were plated in triplicate in semisolid methylcellulose medium (Methocult, STEMCELL Technologies; cat. no. M3434) at 1,000 cells/mL. Colony counts were carried out after 10 days of incubation.
sgRNA Design
sgRNAs were designed using http://cripsr.mit.edu (quality score >70). For every gene, sgRNAs were targeted against RBDs (when annotated) or other protein family (Pfam) domains to maximize negative selection phenotypes (ref. 31; see Supplementary Table S2 for an overview of all sgRNA sequences included in the libraries). sgRNAs were amplified as a pool and cloned into BsmBI (NEB; cat. no. R0580S) digested pLKO1-CRISPR-H2B-GFP. Stbl4 electrocompetent cells (Thermo Fisher Scientific cat. no. 11635018) were transformed, followed by DNA purification from 12 dishes of transformants (PureLink HiPure Plasmid Maxiprep Kit, Thermo Fisher Scientific; cat. no. K210007).
Lentivirus Production
Lentivirus was prepared by transient transfection of 293FT (RRID: CVCL_6911) cells with pMD2.G (RRID: Addgene_12259) and psPAX2 (RRID: Addgene_12260) packaging plasmids to create VSV-G pseudotyped lentiviral particles, as previously described (111). All viral preparations were ultracentrifugated, resuspended in low volumes, and titered on HeLa cells (RRID: CVCL_0030) before being used to infect primary cells and cell lines.
In Vivo Pooled Dropout Screen Transduction and Transplantation
One million tertiary transplant RN2c cells/mouse were expanded in vivo (n = 4), and leukemic BM was harvested from moribund mice and cultured in fresh RPMI supplemented with 10% FBS and 5 μg/mL polybrene (MilliporeSigma; cat. no. H9268). Pretitrated lentivirus (RBP and NTC pools) was added at a clonal MOI of 0.2 with ∼300× coverage, and cultures were incubated overnight, after which they were spun for 5 minutes at 1,200 rpm, resuspended in fresh RPMI supplemented with 10% FBS, and incubated for an additional 24 hours. The H2B-GFP+ fraction was determined at 48 hours after transduction (18%–22%), one-third of the cultures were frozen down for sequencing, and the remaining cells were transplanted into sublethally irradiated B6.SJL (Ly5.1, RRID: IMSR_JAX:002014) recipient mice (2 × 106 cells/mouse, n = 7–9). After 10 days (T10 primary), leukemic BM and splenocytes were isolated and frozen down for sequencing and subsequent analysis (cells from every 2–3 mice were pooled to serve as biological replicates). Secondary mice were transplanted with a portion of the primary mouse BM taken after thawing (T0 Primary) and at the 10-day endpoint (T10 secondary) BM and splenocytes again harvested (samples from every 2–3 mice were pooled). Total DNA from T0, T10 primary, T0 secondary, and T10 secondary cells was isolated using the DNeasy Blood and Tissue Kit (Qiagen; cat. no. 69504) according to the manufacturer's instructions and then further purified using RNeasy columns (Qiagen; cat. no. 74104). sgRNA sequences were then PCR-amplified using Q5 Hot Start High-Fidelity 2X Master Mix (NEB; cat. no. M0494), barcoded primer pairs (see Supplementary Table S2), and 1 μg of DNA per PCR reaction. Individual 50 μL reactions were run on a 3% agarose gel, and libraries were purified using the Zymoclean Gel DNA Recovery kit (Zymo Research; cat. no. D4007). Sequencing was performed using standard Illumina instructions. MAGeCK (ref. 37; https://sourceforge.net/projects/mageck/) was used for the analysis of sequencing reads and calculation of enrichment/depletion of individual sgRNAs. Sufficiency of sgRNA representation of greater than 500 was verified through all arms of the two-step screen.
shRNA Design and qRT-PCR
An Ametrine fluorescent protein was cloned into the pZIP-mCMV-ZsGreen-Puro vector (Transomic Technologies) by amplification of Ametrine from the MNDU3-MLL-AF9-PGK-Ametrine vector (kind gift from Dr. Guy Sauvageau, University of Montreal) with addition of AclI/AgeI restriction sites (NEB; cat. no. R0598S/R3552S) and subsequent subcloning into AclI/AgeI digested pZIP. These ELAVL1 shRNAs (5′-TTGTTAGTGTACAACTCATTT-3′ and 5′-GCAGCATTGGTGAAGTTGAAT-3′) were adapted from the MISSION shRNA library (MilliporeSigma). Annealed oligos were digested with FastDigest XhoI/EcoRI restriction enzymes (Thermo Fisher Scientific; cat. no. FD0694/FD0274) and cloned into the pZIP-mCMV vector. Confirmatory sequencing was carried out for all shRNAs before the generation of lentivirus. Knockdown efficiency of shRNAs was determined by qRT-PCR. For all qRT-PCR determinations total cellular RNA was isolated with TRIzol LS reagent (Thermo Fisher Scientific; cat. no. 10296028) according to the manufacturer's instructions and cDNA was synthesized using the qScript cDNA Synthesis Kit (Quanta Biosciences; cat. no. 95048-100). The mRNA content of samples compared by qRT-PCR was normalized based on the amplification of GAPDH. qRT-PCR was done in triplicate with PerfeCTa qPCR SuperMix Low ROX (Quanta Biosciences; cat. no. 95052-500) with gene-specific probes (Universal Probe Library, UPL, Roche) and primers. See Supplementary Table S2 for all shRNA sequences.
MLL-AF9 and bcCML Infections and Transplants
MLL-AF9 or bcCML cells were thawed and seeded in ultra-low attachment plates at a density of 0.5 × 106 cells/mL of growth media. After 2 to 3 hours of recovery, cells were infected with pZIP-mCMV-Ametrine-shLuciferase (5′-CGATATGGGCTGAATACAAAT-3′), -shElavl1.m1 (5′-CGAGGTTGAATCTGCAAAGCT-3′), or -shElavl1.m2 (5′-CCCACAAATGTTAGACCAATT-3′) at an MOI of 0.5 supplemented with 5 μg/mL Polybrene (MilliporeSigma; cat. no. H9268) at dose equivalents of ∼63,000 and ∼31,000 cells per mouse. Cells were transplanted intravenously 24 hours after infection into sublethally irradiated (580 Rad) C57Blk/6 (Ly5.2, RRID: IMSR_JAX:000664) recipient mice. BM and spleens were harvested 14 and 9 days after transplant for the MLL-AF9 and bcCML recipients, respectively, for flow-cytometric analysis.
Isolation of Mouse Stem and Progenitor Cell Populations
BM and spleen were harvested from 6- to 12-week-old mice, crushed in IMDM + 3% FBS and passed through 40-μm cell strainers. Ammonium chloride (STEMCELL Technologies; cat. no. 07850) was used for lysis of red blood cells, followed by incubation of the cells with a cocktail of lineage-specific antibodies (CD5, CD11b, CD19, B220, Gr-1, and TER119; STEMCELL Technologies; cat. no. 19856) for negative selection of Lin− cells using an EasySep immunomagnetic column (STEMCELL Technologies; cat. no. 18000). Live cells were discriminated on the basis of cell size, granularity and, as needed, absence of viability dye 7-AAD (BD Biosciences; cat. no. 559925, RRID: AB_2869266) uptake.
Competitive Mouse HSPC Transplants
Freshly sorted mouse Lin−CD150+CD48− BM cells were seeded at 3,000 cells per well in 96-well plates and cultured in SFEM (STEMCELL Technologies; cat. no. 09650) supplemented with 10 ng/mL mouse IL3 (R&D Systems; cat. no. 403-ML-050), 10 ng/mL mouse IL6 (R&D Systems; cat. no. 406-ML-025), 100 ng/mL mouse SCF (R&D Systems; cat. no. 455-MC-010), and 100 ng/mL mouse TPO (PeproTech; cat. no. AF-315-14). Cultures were prestimulated for 24 hours, followed by lentiviral infection with shRNAs targeting Elavl1 and Luciferase (pZIP-mEF1a-ZsGreen-miR-E) in 8 μg/mL Polybrene (MilliporeSigma; cat. no. H9268). Percent of gene transfer was determined 72 hours after transduction, followed by transplantation of the cultures (1/4 of each well per mouse, n = 4) along with 1 × 105 whole BM competitor cells (C57Blk/6, Ly5.2+, RRID: IMSR_JAX:000664). Donor-derived reconstitution was determined at 4-week intervals by examining peripheral blood samples from each mouse for Ly5.1+ZsGreen+ fractions; BM and spleens were harvested from all recipients 18 weeks after transplant for immunophenotyping. For MLL-AF9 and bcCML cell lines, leukemic BM and splenocytes were freshly harvested from primary transplanted, moribund mice, followed by 24 to 48 hours lentiviral transductions with pretitrated virus (pZIP-mCMV-Ametrine-shLuciferase/ and -shElavl1) in the presence of 5 μg/mL Polybrene (MilliporeSigma; cat. no. H9268). On the day of the transplant, the percentage of gene transfer was determined by flow cytometry, and 100,000 cells were intravenously transplanted into sublethally irradiated (580 Rad) C57Blk/6 (RRID: IMSR_JAX:000664) recipient mice. The Ametrine+ fraction of the leukemic grafts was flow cytometrically analyzed from BM and spleens of moribund mice and compared with the Ametrine+ fraction on the day of transplant to determine the effect of KD on in vivo leukemic propagation.
Human BM Cell Subset RNA-seq and Analysis
Human adult BM samples were obtained with written informed consent and conducted in accordance with recognized ethical guidelines by the Research Ethics Board at the UHN (REB # 01-0573-C) in accordance with Canadian Tri-Council Policy Statement on the Ethical Conduct for Research Involving Humans (TCPS). Hematopoietic stem and progenitors from patient BM samples were sorted using a BD FACSAria (BD Biosciences). 4–5 × 103 HSCs and progenitors were sorted from at least 3 BM samples with the following phenotypes: LT-HSC, CD34+CD38−CD90+CD49f+; ST-HSC, CD34+CD38−CD90+CD49f−; MPP, multipotent progenitor (CD34+CD38−CD45RA−CD90−CD49f−). RNA from 2–4 × 103 cells was isolated using the PicoPure RNA Isolation Kit (Thermo Fisher Scientific; cat. no. KIT0204). cDNA was generated and amplified using the SMART-SEQ2 method (112). Sequencing libraries were generated with the low input RNA Nextera protocol (Nextera DNA Sample Preparation Kit, Illumina; cat. no. FC-121-1031). Samples sequenced were subjected to 125 base-pair (bp), paired-end RNA-seq on the Illumina HiSeq 2500 with V4 flowcells, with an average of 50 million reads/sample. RNA-seq data were mapped using STAR aligner (RRID: SCR_004463; ref. 113) against the hg38 reference genome. The FPKM and expression values were calculated using the Tuxedo suits tool kit (114), and R was used for downstream statistical analysis.
Enhanced Cross-linking and Immunoprecipitation
bcCML cells were freshly harvested, crushed in IMDM + 3% FBS, and passed through 40-μm cell strainers followed by red blood cell lysis with Ammonium Chloride (STEMCELL Technologies; cat. no. 07850). Cells were incubated with a cocktail of lineage-specific antibodies (CD5, CD11b, CD19, B220, Gr-1, and TER119; STEMCELL Technologies; cat. no. 19856) for negative selection of Lin− cells using an EasySep immunomagnetic column (STEMCELL Technologies; cat. no. 18000). 20 × 106 cells per sample were subsequently washed in PBS and UV-crosslinked at 400 mJ/cm2 on ice. Samples were then pelleted, snap-frozen, and stored at −80°C. Enhanced Cross-linking and Immunoprecipitation (eCLIP) sequencing was performed as previously described (67). Pellets were lysed in iCLIP lysis buffer, treated with RNase I (NEB; cat. no. M0314L) for 5 minutes at 37°C, followed by immunoprecipitation using 10 μg anti-ELAVL1 mouse monoclonal IgG (Abcam; cat. no. ab170193) and 125 μL M-280 Sheep-α-Mouse IgG Dynabeads (Thermo Fisher Scientific; cat. no. 11201D) per sample. After stringent rounds of washing, samples were dephosphorylated (FastAP, Thermo Fisher Scientific; cat. no. EF0654; T4 PNK, NEB; cat. no. M0201L) and 3′ ligation (on-bead) with a barcoded RNA adapter followed using T4 RNA Ligase (NEB; cat. no. M0437M). Samples were again stringently washed, run on standard protein gels, and transferred to nitrocellulose membranes. The region spanning 36–115 kDa was then isolated, followed by extraction of RNA from the membranes, and reverse transcribed (AffinityScript; Agilent). A DNA adapter containing a 5′ random-mer was then ligated (3′), followed by the cleanup of the samples and PCR amplification. Libraries were size-selected (175–350 bp) and purified from 3% low melting temperature agarose gels, followed by sequencing on the Illumina HiSeq 2500 platform (paired-end, 50 nt). Reads were processed using the eCLIP processing pipeline for paired-end data, summarized as follows. Briefly, reads were demultiplexed according to their inline barcodes (A01, B06 for rep1, C01, D8f for rep2) and 10 nt unique molecular identifiers (UMI) were extracted. Reads were then trimmed twice with Cutadapt (v1.14.0) to remove adapters and adapter dimer sequences. Reads were then aligned to mouse genome (mm9) with STAR (v2.4.0i), selecting for uniquely mapped reads (--outFilterMultimapNmax 1). Uniquely mapped reads were then sorted and collapsed according to their UMI tags to remove PCR duplicates. These reads were then merged according to their inline barcodes, yielding 3,532,702 and 4,027,997 usable reads in the two ELAVL1 immunoprecipitation (IP) replicates. Corresponding size-matched input controls were processed the same way and found 1,525,778 and 1,394,954 usable reads. Peaks were then called and normalized with CLIPper (available at: https://github.com/YeoLab/clipper) and with custom scripts. These scripts, as well as all pipeline definitions and details, are available at: https://github.com/YeloLab/eCLIP. After identifying peaks enriched above input control [−log10(P value) ≥ 3 and log2(fold) ≥ 3] and performing irreproducibility discovery rate analysis (67), 4,346 reproducible peaks were found. Downstream analysis was performed using BEDTools (RRID: SCR_006646), HOMER (RRID: SCR_010881), and Pathway Analysis. See Supplementary Table S2 for RNA and DNA adapters used.
RNA-seq of RN2c Cells
RN2c cells were transduced with lentiviral-packaged sgRNA targeting Elavl1 and Ano9 as control. Forty-eight hours following transduction, live 7AAD− GFP+ cells were isolated by FACS, and RNA was extracted by TRIzol LS (Thermo Fisher Scientific; cat. no. 10296028) following the manufacturer's protocol. RNA was coprecipitated with GlycoBlue (Thermo Fisher Scientific; cat. no. AM9516) following the manufacturer's protocol; in brief, 3 μL (final concentration 45 μg/mL) of GlycoBlue was added to TRIzol LS-separated aqueous phase followed by one volume isopropanol and incubated at −20°C for 30 minutes before pelleting RNA by centrifugation. Isolated RNA was treated with DNAse I (Thermo Fisher Scientific; cat. no. EN0521) which was inactivated by the addition of EDTA and incubation at 65°C. PolyA+ RNA libraries were generated using NEBNext PolyA mRNA Magnetic Isolation Module (NEB; cat. no. E7490) and NEBNext Ultra II Directional RNA Library Prep Kit for Illumina (NEB; cat. no. E7760L) and indexed using NEBNext Multiplex Oligos for Illumina (NEB; cat. no. E6440L). 100 bp paired-end sequencing was performed on the Illumina HiSeq 1500 at a depth of 50 million reads per sample. Sequencing reads were processed to identify differential gene expression via Yeo lab's in-house pipeline that performs the following steps: First, reads were trimmed of standard illumine adapters with Cutadapt (v1.14.0) and checked for quality with fastQC (v0.10.1). Trimmed reads were mapped first to RepBase (18.05) with STAR (v2.4.0), then mapped to the mm9 genome using end-to-end alignment (–alignEndsType EndToEnd). Aligned reads were annotated with featureCounts (v1.5.0-p1) using Gencode vM1 annotations and differential expression was performed with DESeq2 (v1.14.1, RRID: SCR_000154). AS was determined using rMATS (v3.2.5), and significant nonoverlapping alternative splice events were assigned where FDR <0.05 and lncLevelDifference was magnitude 0.1 or higher.
RNA-seq of Primary AML Cells
Human primary AML cells were infected in triplicate with pLKO.1-EGFP-shScramble or -shELAVL1.2 (5′-CCCATCACAGTGAAGTTTGCA-3′) at an MOI of 50 for 48 hours, after which 7AAD-EGFP+ cells were sorted, and 100,000 cells per condition were resuspended in TRIzol LS (Thermo Fisher Scientific; cat. no. 10296028) following the manufacturer's protocol. The aqueous phase was separated using Phasemaker Tubes (Thermo Fisher Scientific; cat. no. A33248). Isolated RNA was treated with DNAse from TURBO DNA-free kit (Thermo Fisher Scientific; cat. no. AM1907). 100 bp paired-end sequencing was performed on the Illumina HiSeq 1500 at a depth of 50 million reads per sample. Sequencing reads were checked for quality with fastQC (v0.10.1) and aligned to the hg38 reference genome using STAR (v2.7.2c). Quantification and differential expression were performed using RSEM (v1.3.1) and DESeq2 (v1.26.0). AS was determined using rMATS (v4.0.2) and significant nonoverlapping alternative splice events were assigned where FDR <0.05 and lncLevelDifference was magnitude 0.1 or higher.
Expression Profiling Analysis of AML Patient Diagnosis–Relapse Paired Samples
ELAVL1 expression values were normalized in logTPM from three publicly available data sets (3, 55, 56), and a Wilcoxon signed-rank test was used to compare the groups. We included only pairs in which the relapse followed chemotherapy treatment.
Gene Set and Pathway Analysis
GO annotations were assigned to screening candidates and hits using the open-source web application GOnet (Pomaznoy M, Ha B, Peters B. GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinformatics. 2018;19:470). Above- and below-median cohorts for ELAVL1 expression were identified in the BeatAML RNA-seq data set (78) using DESeq-normalized counts, and altered genesets were identified using DESeq2 followed by fGSEA. The fgsea R package (v1.12.0) was used to perform preranked GSEA against a gene set repository maintained by Dr. Gary Bader (Princess Margaret Cancer Centre), which encompasses gene sets from GO Biological Processes, Reactome, and MSigDB. Rank scores were calculated as −log10(P)*sign(log2FC). Enrichments were visualized using the Cytoscape Enrichment Map plugin. Pathway analyses for differentially spliced genes and eCLIP targets were performed using g:Profiler.
Flow Cytometry
All flow cytometry analysis was performed using a MACSQuant Analyzer 10 (Miltenyi Biotec), BD LSRII Analyzer, BD LSRFortessa, or BD LSRFortessa X-20 (BD Biosciences). Analysis was performed using FlowJo software (Tree Star, RRID: SCR_008520).
MitoTracker Experiments
Primary AML cells were incubated with 2 μmol/L FCCP (Cayman Chemicals; cat. no. 15218) for 30 minutes at 37°C followed by 2 μmol/L FCCP with 50 nmol/L MitoTracker Orange CMTMRos (MTO; Thermo Fisher Scientific # M7510) for 30 minutes at 37°C for 72 hours after infection or after treatment with 5 μmol/L MS-444 (MedChemExpress; cat. no. HY-100685). RN2c cells were incubated with 100 nmol/L MTO (Thermo Fisher Scientific # M7510) for 30 minutes at 37°C for 48 hours after infection. Primary AML cells were stained with 7-AAD (BD Biosciences; cat. no. 559925, RRID: AB_2869266) and RN2c cells were stained with SYTOX Blue Dead Cell Stain (Thermo Fisher Scientific; cat. no. S34857) at their corresponding time points. The median fluorescence intensity of MTO in live cells was quantified by flow cytometry.
Western Blot
Immunoblotting was performed with anti-ELAVL1 mouse monoclonal IgG (Abcam; cat. no. ab170193), anti-RBM14 rabbit polyclonal (Abcam; cat. no. ab70636), anti-TOMM34 rabbit polyclonal (Abcam; cat. no. ab230103), anti-β-actin mouse monoclonal (MilliporeSigma; cat. no. A5441), and α-Tubulin rabbit monoclonal IgG (Cell Signaling Technologies; cat. no. 2125S) antibodies. Secondary antibodies used were IRDye 680RD goat anti-rabbit IgG (LI-COR Biosciences; cat. no. 926-68071, RRID: AB_10956166), IRDye 680RD goat anti-mouse IgG (LI-COR Biosciences; cat. no. 925-68070, RRID: AB_2651128), and IRDye 800CW goat anti-mouse IgG (LI-COR Biosciences; cat. no. 926-32210, RRID: AB_621842).
Immunofluorescence
Approximately 180,000 primary AML or RN2c cells were prepared for staining by cytocentrifugation (600 rpm, 5 minutes). Cells were fixed in 4% PFA (Electron Microscopy Sciences; cat. no. 15710) for 20 minutes, followed by permeabilization in 0.1× Triton X-100 (Bioshop; cat. no. TRX777) in PBS for 10 minutes. Samples were incubated in blocking buffer (PBST + 10% goat serum + 1% W/V BSA) for 1 hours at room temperature, followed by incubation with anti-ELAVL1 (Abcam; cat. no. ab170193) and anti-G3BP1 (Proteintech; cat. no. 13057-2-AP) overnight at 4°C. Secondary antibody incubation was performed with AlexaFluor-647 donkey-anti-mouse IgG (Thermo Fisher Scientific; cat. no. A21235) or AlexaFluor-488 donkey-anti-rabbit IgG (Thermo Fisher Scientific; cat. no. A21206) for 1 hour at room temperature with DAPI staining (Thermo Fisher Scientific; cat. no. D21490) performed simultaneously. Slides were mounted with Fluoromount mounting medium (Thermo Fisher Scientific; cat. no. 00-4958-02) and images were captured using an Olympus IX81 microscope (40× objective lens).
Statistical Analysis
Unless stated otherwise (i.e., analysis of RNA-seq data sets), all statistical analysis was performed using GraphPad Prism (GraphPad Software version 5.0, RRID: SCR_002798). Unpaired Student t tests were performed with P < 0.05 as the cutoff for statistical significance. CRISPR scores from (41) were analyzed using MATLAB (R2014B, The MathWorks Inc, RRID: SCR_001622).
Data Availability
The eCLIP and mouse RNA-seq data generated in this study were deposited and are publicly available in Gene-Expression Omnibus (GEO) at accession references GSE127944 and GSE127743, respectively. Human AML RNA-seq data set: GSE224548. All other data supporting the findings of this study are cited in Materials and Methods, in supplementary documents, or are available upon request from the authors.
Supplementary Material
Table S1 shows expression of RBPs in an RNA-sequencing data set of human bone marrow hematopoietic stem and progenitor subpopulations.
Table S2 shows sequences of (a) sgRNAs from the RBP screen, (b) sequencing primers, (c) shRNAs and human-targeting sgRNAs, (d) eCLIP oligos.
Table S3 shows clinical information for all AML patient specimens used in this study.
Table S4 shows limiting dilution analysis of secondary recipients transplanted with DMSO- or MS-444-treated bone marrow from primary mice
Table S5 shows the log2 fold change of differentially expressed transcripts from the ELAVL1 knockout RN2c RNA-sequencing data set.
Table S6 shows altered gene sets identified by GSEA of the ELAVL1 knockout RN2c RNA-sequencing data set.
Table S7 shows (a) reproducible peaks and (b) GO analysis of ELAVL1-bound transcripts identified in the eCLIP-sequencing data set. The negatively enriched (c) and positively-enriched (d) gene sets identified by GSEA of ELAVL1-bound and differentially expressed transcripts from the RN2c RNA-sequencing data set are shown.
Table S8 shows transcripts from the ELAVL1 knockout RNA-sequencing data set that are (a) alternatively spliced and (b) alternatively spliced as well as bound by ELAVL1 as identified by eCLIP-sequencing.
Table S9 shows the (a) differentially expressed transcripts and (b) altered pathways as identified by GSEA from the ELAVL1 knockdown human primary AML RNA-sequencing data set.
Supplemental Figure S1 shows expression of RBPs in healthy HSPCs and RN2c cells, sgRNA drop out dynamics in primary and secondary transplants, the molecular functions and biological processes associated with the hit RBPs and their gene essentialities.
Supplemental Figure S2 shows validation of sgRNA -mediated knockout of hit RBPs, comparison of hit RBP depletion in RN2c in vitro versus in vivo, effects of ELAVL1 loss on human AML cell lines, characteristics of the MLL-AF9 AML mouse model, and ELAVL1 expression in normal hematopoietic cells.
Supplemental Figure S3 shows the generation of the bcCML mouse model and the effects of ELAVL1 on normal bone marrow in vivo.
Supplemental Figure S4 shows the validation of ELAVL1 knockdown by western blot and effects of ELAVL1 loss on human primary AML cells in vitro and in vivo.
Supplemental Figure S5 shows the effects of DHTS treatment on THP-1 and human primary AML cells in vitro, the effects of MS-444 treatment on human umbilical cord blood in vivo, and validation of ELAVL1 overexpression by western blot.
Supplemental Figure S6 shows the GSEA of ELAVL1 depletion in mouse RN2c and human primary AML cells, ELAVL1 subcellular localization in mouse and human AML, the qRT-PCR of ELAVL1 targets in MS-444-treated RN2c cells, and the integrative analysis of the mouse RNA-seq and eCLIP-seq.
Supplemental Figure S7 shows the expression profile of TOMM34 in normal human bone marrow and across different AML and CML data sets. The effects of TOMM34 depletion on human primary AML cells in vitro and validation of TOMM34 knockdown and overexpression is shown.
Acknowledgments
The authors thank Brad Doble, Bernardino Trigatti, Jon Draper, and all members of the Hope lab for important feedback on this work; Minomi Subapanditha and Zoya Shapovalova for flow cytometry sorting; and Lillian Robson, Wendy Whittaker, and Norma-Ann Kearns for animal caretaking and maintenance. This work was supported by a Prins Bernhard Cultuurfonds Fellowship (L. de Rooij), a Canadian Cancer Society (CCS) Post-doctoral Fellowship (S.K. Loganathan), a Canadian Institutes of Health Research (CIHR) MD/PhD Studentship (D.C. Chan, H.T. Chen), an Ontario Graduate Scholarship (D.C. Chan and A. Vujovic), a CIHR Doctoral Fellowship (A. Keyvani Chahi and J. Xu), a CIHR Canada Graduate Scholarship-Master's Award (A. Tajik, E. Tsao, and P. Joshi), an Ontario Institute for Cancer Research (OICR) Acute Leukemia Translational Research Initiative Grant (J.E. Dick and K.J. Hope), CIHR Grant (#RN380110-409786), CCS Grant (#703212), a CIHR Project Grant (#438792; D. Schramek), an NIH R01 Grant (#HL137223; G.W. Yeo and K.J. Hope) and an OICR Investigator Award (K.J. Hope).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
S.K. Loganathan reports grants from the Canadian Cancer Society during the conduct of the study. P. Joshi reports other support from Canadian Institutes of Health Research during the conduct of the study. J.E. Dick reports grants from CIHR, CCSRI, and OICR during the conduct of the study; other support from Celgene/BMS outside the submitted work; in addition, J.E. Dick has a patent for SIRP-A targeting issued and licensed to Trillium Therapeutics/Pfizer. D. Schramek reports personal fees from Tango Therapeutics outside the submitted work. G.W. Yeo reports grants from NIH during the conduct of the study. K.J. Hope reports grants from the Canadian Institutes of Health Research, Ontario Institutes for Health Research, and NIH during the conduct of the study; other support from Twentyeight-seven Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
A. Vujovic: Conceptualization, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. de Rooij: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A. Keyvani Chahi: Conceptualization, data curation, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. H.T. Chen: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. B.A. Yee: Data curation, software, formal analysis, investigation, methodology, writing–review and editing. S.K. Loganathan: Conceptualization, resources, data curation, software, formal analysis, investigation, methodology, writing–review and editing. L. Liu: Conceptualization, resources, data curation, formal analysis, investigation, methodology, writing–review and editing. D.C.H. Chan: Formal analysis, validation, investigation, methodology, writing–review and editing. A. Tajik: Formal analysis, validation, investigation, writing–review and editing. E. Tsao: Formal analysis, investigation, writing–review and editing. S. Moreira: Investigation, writing–review and editing. P. Joshi: Investigation, writing–review and editing. J. Xu: Investigation, writing–review and editing. N. Wong: Investigation, project administration, writing–review and editing. Z. Balde: Resources, data curation, investigation, methodology, project administration, writing–review and editing. S. Jahangiri: Resources, data curation, formal analysis, supervision, investigation, writing–review and editing. S. Zandi: Resources, data curation, supervision, investigation, project administration, writing–review and editing. S. Aigner: Resources, data curation, supervision, investigation, writing–review and editing. J.E. Dick: Resources, supervision, investigation, methodology, writing–review and editing. M.D. Minden: Resources, software, supervision, funding acquisition, methodology, writing–review and editing. D. Schramek: Conceptualization, resources, software, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing. G.W. Yeo: Resources, software, supervision, funding acquisition, methodology, writing–review and editing. K.J. Hope: Conceptualization, resources, data curation, software, formal analysis, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373:1136–52. [DOI] [PubMed] [Google Scholar]
- 2. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275–91. [DOI] [PubMed] [Google Scholar]
- 3. Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 2017;547:104–8. [DOI] [PubMed] [Google Scholar]
- 4. Guzman ML, Neering SJ, Upchurch D, Grimes B, Howard DS, Rizzieri DA, et al. Nuclear factor-kappaB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 2001;98:2301–7. [DOI] [PubMed] [Google Scholar]
- 5. Guan Y, Gerhard B, Hogge DE. Detection, isolation, and stimulation of quiescent primitive leukemic progenitor cells from patients with acute myeloid leukemia (AML). Blood 2003;101:3142–9. [DOI] [PubMed] [Google Scholar]
- 6. Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013;12:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ishikawa F, Yoshida S, Saito Y, Hijikata A, Kitamura H, Tanaka S, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol 2007;25:1315–21. [DOI] [PubMed] [Google Scholar]
- 8. Ye H, Adane B, Khan N, Sullivan T, Minhajuddin M, Gasparetto M, et al. Leukemic stem cells evade chemotherapy by metabolic adaptation to an adipose tissue niche. Cell Stem Cell 2016;19:23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamauchi T, Masuda T, Canver MC, Seiler M, Semba Y, Shboul M, et al. Genome-wide CRISPR-Cas9 screen identifies leukemia-specific dependence on a pre-mRNA metabolic pathway regulated by DCPS. Cancer Cell 2018;33:386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bajaj J, Hamilton M, Shima Y, Chambers K, Spinler K, Van Nostrand EL, et al. An in vivo genome-wide CRISPR screen identifies the RNA-binding protein Staufen2 as a key regulator of myeloid leukemia. Nat Cancer 2020;1:410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin S, Larrue C, Scheidegger NK, Seong BKA, Dharia NV, Kuljanin M, et al. An in vivo CRISPR screening platform for prioritizing therapeutic targets in AML. Cancer Discov 2022;12:432–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 2007;8:533–43. [DOI] [PubMed] [Google Scholar]
- 13. Neelamraju Y, Gonzalez-Perez A, Bhat-Nakshatri P, Nakshatri H, Janga SC. Mutational landscape of RNA-binding proteins in human cancers. RNA Biol 2018;15:115–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, et al. ADAR1 activation drives leukemia stem cell self-renewal by impairing let-7 biogenesis. Cell Stem Cell 2016;19:177–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the RNA m(6)A reader YTHDF2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell 2019;25:137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-binding protein network in acute myeloid leukemia. Cancer Cell 2019;35:369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang J, Li Y, Wang P, Han G, Zhang T, Chang J, et al. Leukemogenic chromatin alterations promote AML leukemia stem cells via a KDM4C-ALKBH5-AXL signaling axis. Cell Stem Cell 2020;27:81–97. [DOI] [PubMed] [Google Scholar]
- 18. Park SM, Gonen M, Vu L, Minuesa G, Tivnan P, Barlowe TS, et al. Musashi2 sustains the mixed-lineage leukemia-driven stem cell regulatory program. J Clin Invest 2015;125:1286–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rentas S, Holzapfel N, Belew MS, Pratt G, Voisin V, Wilhelm BT, et al. Musashi-2 attenuates AHR signalling to expand human haematopoietic stem cells. Nature 2016;532:508–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, et al. METTL14 inhibits hematopoietic stem/progenitor differentiation and promotes leukemogenesis via mRNA m(6)A modification. Cell Stem Cell 2018;22:191–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, et al. The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat Med 2017;23:1369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cheng F, Pan Y, Lu YM, Zhu L, Chen S. RNA-binding protein Dnd1 promotes breast cancer apoptosis by stabilizing the bim mRNA in a miR-221 binding site. Biomed Res Int 2017;2017:9596152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiou GY, Yang TW, Huang CC, Tang CY, Yen JY, Tsai MC, et al. Musashi-1 promotes a cancer stem cell lineage and chemoresistance in colorectal cancer cells. Sci Rep 2017;7:2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Degrauwe N, Suva ML, Janiszewska M, Riggi N, Stamenkovic I. IMPs: an RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev 2016;30:2459–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kechavarzi B, Janga SC. Dissecting the expression landscape of RNA-binding proteins in human cancers. Genome Biol 2014;15:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mongroo PS, Noubissi FK, Cuatrecasas M, Kalabis J, King CE, Johnstone CN, et al. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res 2011;71:2172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet 2014;15:829–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 2016;540:433–7. [DOI] [PubMed] [Google Scholar]
- 29. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med 2011;17:1086–93. [DOI] [PubMed] [Google Scholar]
- 30. Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature 2006;442:818–22. [DOI] [PubMed] [Google Scholar]
- 31. Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, Vakoc CR. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotechnol 2015;33:661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Somervaille TC, Cleary ML. Identification and characterization of leukemia stem cells in murine MLL-AF9 acute myeloid leukemia. Cancer Cell 2006;10:257–68. [DOI] [PubMed] [Google Scholar]
- 33. Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller PG, Al-Shahrour F, Hartwell KA, Chu LP, Jaras M, Puram RV, et al. In Vivo RNAi screening identifies a leukemia-specific dependence on integrin beta 3 signaling. Cancer Cell 2013;24:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Puram RV, Kowalczyk MS, de Boer CG, Schneider RK, Miller PG, McConkey M, et al. Core circadian clock genes regulate leukemia stem cells in AML. Cell 2016;165:303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Loganathan SK, Schleicher K, Malik A, Quevedo R, Langille E, Teng K, et al. Rare driver mutations in head and neck squamous cell carcinomas converge on NOTCH signaling. Science 2020;367:1264–9. [DOI] [PubMed] [Google Scholar]
- 37. Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 2014;15:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Walker CJ, Oaks JJ, Santhanam R, Neviani P, Harb JG, Ferenchak G, et al. Preclinical and clinical efficacy of XPO1/CRM1 inhibition by the karyopherin inhibitor KPT-330 in Ph+ leukemias. Blood 2013;122:3034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Etchin J, Berezovskaya A, Conway AS, Galinsky IA, Stone RM, Baloglu E, et al. KPT-8602, a second-generation inhibitor of XPO1-mediated nuclear export, is well tolerated and highly active against AML blasts and leukemia-initiating cells. Leukemia 2017;31:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naudin C, Hattabi A, Michelet F, Miri-Nezhad A, Benyoucef A, Pflumio F, et al. PUMILIO/FOXP1 signaling drives expansion of hematopoietic stem/progenitor and leukemia cells. Blood 2017;129:2493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang T, Yu H, Hughes NW, Liu B, Kendirli A, Klein K, et al. Gene essentiality profiling reveals gene networks and synthetic lethal interactions with oncogenic ras. Cell 2017;168:890–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Conant D, Hsiau T, Rossi N, Oki J, Maures T, Waite K, et al. Inference of CRISPR edits from sanger trace data. CRISPR J 2022;5:123–30. [DOI] [PubMed] [Google Scholar]
- 43. Klimmeck D, Cabezas-Wallscheid N, Reyes A, von Paleske L, Renders S, Hansson J, et al. Transcriptome-wide profiling and posttranscriptional analysis of hematopoietic stem/progenitor cell differentiation toward myeloid commitment. Stem cell reports 2014;3:858–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bagger FO, Sasivarevic D, Sohi SH, Laursen LG, Pundhir S, Sonderby CK, et al. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res 2016;44:D917–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res 2002;12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, et al. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Natl Acad Sci U S A. 2006;103:2794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schoof EM, Furtwangler B, Uresin N, Rapin N, Savickas S, Gentil C, et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat Commun 2021;12:3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rapin N, Bagger FO, Jendholm J, Mora-Jensen H, Krogh A, Kohlmann A, et al. Comparing cancer vs normal gene expression profiles identifies new disease entities and common transcriptional programs in AML patients. Blood 2014;123:894–904. [DOI] [PubMed] [Google Scholar]
- 49. Kohlmann A, Kipps TJ, Rassenti LZ, Downing JR, Shurtleff SA, Mills KI, et al. An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: the Microarray Innovations in LEukemia study prephase. Br J Haematol 2008;142:802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Klein HU, Ruckert C, Kohlmann A, Bullinger L, Thiede C, Haferlach T, et al. Quantitative comparison of microarray experiments with published leukemia related gene expression signatures. BMC Bioinf 2009;10:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Metzelder SK, Michel C, von Bonin M, Rehberger M, Hessmann E, Inselmann S, et al. NFATc1 as a therapeutic target in FLT3-ITD-positive AML. Leukemia 2015;29:1470–7. [DOI] [PubMed] [Google Scholar]
- 52. Wouters BJ, Lowenberg B, Erpelinck-Verschueren CA, van Putten WL, Valk PJ, Delwel R. Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 2009;113:3088–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cancer Genome Atlas Research N, Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Metzeler KH, Hummel M, Bloomfield CD, Spiekermann K, Braess J, Sauerland MC, et al. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood 2008;112:4193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li S, Garrett-Bakelman FE, Chung SS, Sanders MA, Hricik T, Rapaport F, et al. Distinct evolution and dynamics of epigenetic and genetic heterogeneity in acute myeloid leukemia. Nat Med 2016;22:792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, et al. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci U S A 2002;99:7622–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neering SJ, Bushnell T, Sozer S, Ashton J, Rossi RM, Wang PY, et al. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood 2007;110:2578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ashton JM, Balys M, Neering SJ, Hassane DC, Cowley G, Root DE, et al. Gene sets identified with oncogene cooperativity analysis regulate in vivo growth and survival of leukemia stem cells. Cell Stem Cell 2012;11:359–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, et al. Essential role of the RNA-binding protein HuR in progenitor cell survival in mice. J Clin Invest 2009;119:3530–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tang H, Wang H, Cheng X, Fan X, Yang F, Zhang M, et al. HuR regulates telomerase activity through TERC methylation. Nat Commun 2018;9:2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yilmaz OH, Kiel MJ, Morrison SJ. SLAM family markers are conserved among hematopoietic stem cells from old and reconstituted mice and markedly increase their purity. Blood 2006;107:924–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. D'Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, et al. Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function. Sci Rep 2015;5:16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, et al. Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol 2007;3:508–15. [DOI] [PubMed] [Google Scholar]
- 65. Romeo C, Weber MC, Zarei M, DeCicco D, Chand SN, Lobo AD, et al. HuR contributes to TRAIL resistance by restricting death receptor 4 expression in pancreatic cancer cells. Mol Cancer Res 2016;14:599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lang M, Berry D, Passecker K, Mesteri I, Bhuju S, Ebner F, et al. HuR small-molecule inhibitor elicits differential effects in adenomatosis polyposis and colorectal carcinogenesis. Cancer Res 2017;77:2424–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 2016;13:508–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A. 2004;101:2987–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mukherjee N, Corcoran DL, Nusbaum JD, Reid DW, Georgiev S, Hafner M, et al. Integrative regulatory mapping indicates that the RNA-binding protein HuR couples pre-mRNA processing and mRNA stability. Mol Cell 2011;43:327–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pabst C, Bergeron A, Lavallee VP, Yeh J, Gendron P, Norddahl GL, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 2016;127:2018–27. [DOI] [PubMed] [Google Scholar]
- 71. Choudhury R, Roy SG, Tsai YS, Tripathy A, Graves LM, Wang Z. The splicing activator DAZAP1 integrates splicing control into MEK/Erk-regulated cell proliferation and migration. Nat Commun 2014;5:3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 2018;553:515–20. [DOI] [PubMed] [Google Scholar]
- 73. Liu L, Rao JN, Zou T, Xiao L, Wang PY, Turner DJ, et al. Polyamines regulate c-Myc translation through Chk2-dependent HuR phosphorylation. Mol Biol Cell 2009;20:4885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Perrone EE, Liu L, Turner DJ, Strauch ED. Bile salts increase epithelial cell proliferation through HuR-induced c-Myc expression. J Surg Res 2012;178:155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Liu L, Ouyang M, Rao JN, Zou T, Xiao L, Chung HK, et al. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell 2015;26:1797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li S, Li J, Chen C, Zhang R, Wang K. Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis 2018;5:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zeng C, Liu S, Lu S, Yu X, Lai J, Wu Y, et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Cancer 2018;17:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Egan G, Khan DH, Lee JB, Mirali S, Zhang L, Schimmer AD. Mitochondrial and metabolic pathways regulate nuclear gene expression to control differentiation, stem cell function, and immune response in leukemia. Cancer Discov 2021;11:1052–66. [DOI] [PubMed] [Google Scholar]
- 80. Trcka F, Durech M, Man P, Hernychova L, Muller P, Vojtesek B. The assembly and intermolecular properties of the Hsp70-Tomm34-Hsp90 molecular chaperone complex. J Biol Chem 2014;289:9887–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Trcka F, Durech M, Vankova P, Vandova V, Simoncik O, Kavan D, et al. The interaction of the mitochondrial protein importer TOMM34 with HSP70 is regulated by TOMM34 phosphorylation and binding to 14–3-3 adaptors. J Biol Chem 2020;295:8928–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Durech M, Trcka F, Man P, Blackburn EA, Hernychova L, Dvorakova P, et al. Novel entropically driven conformation-specific interactions with Tomm34 protein modulate Hsp70 protein folding and ATPase activities. Mol Cell Proteomics 2016;15:1710–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang T, Nie Y, Gu J, Cai K, Chen X, Li H, et al. Identification of mitochondrial-related prognostic biomarkers associated with primary bile acid biosynthesis and tumor microenvironment of hepatocellular carcinoma. Front Oncol 2021;11:587479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014;513:382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zhou F, Liu Y, Rohde C, Pauli C, Gerloff D, Kohn M, et al. AML1-ETO requires enhanced C/D box snoRNA/RNP formation to induce self-renewal and leukaemia. Nat Cell Biol 2017;19:844–55. [DOI] [PubMed] [Google Scholar]
- 86. Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003;22:7146–54. [DOI] [PubMed] [Google Scholar]
- 87. Heinonen M, Bono P, Narko K, Chang SH, Lundin J, Joensuu H, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res 2005;65:2157–61. [DOI] [PubMed] [Google Scholar]
- 88. Melling N, Taskin B, Hube-Magg C, Kluth M, Minner S, Koop C, et al. Cytoplasmic accumulation of ELAVL1 is an independent predictor of biochemical recurrence associated with genomic instability in prostate cancer. Prostate 2016;76:259–72. [DOI] [PubMed] [Google Scholar]
- 89. Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16–1 cluster functions in human leukemia. Proc Natl Acad Sci U S A 2008;105:5166–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Topisirovic I, Siddiqui N, Orolicki S, Skrabanek LA, Tremblay M, Hoang T, et al. Stability of eukaryotic translation initiation factor 4E mRNA is regulated by HuR, and this activity is dysregulated in cancer. Mol Cell Biol 2009;29:1152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Scherrer T, Mittal N, Janga SC, Gerber AP. A screen for RNA-binding proteins in yeast indicates dual functions for many enzymes. PLoS One 2010;5:e15499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Castello A, Hentze MW, Preiss T. Metabolic enzymes enjoying new partnerships as RNA-binding proteins. Trends Endocrinol Metab 2015;26:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bakheet T, Hitti E, Al-Saif M, Moghrabi WN, Khabar KSA. The AU-rich element landscape across human transcriptome reveals a large proportion in introns and regulation by ELAVL1/HuR. Biochim Biophys Acta Gene Regul Mech 2018;1861:167–77. [DOI] [PubMed] [Google Scholar]
- 94. Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J 1998;17:3448–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J 1998;17:3461–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3' untranslated regions of cytokine and angiogenic factor mRNAs. Cancer Res 2001;61:2154–61. [PubMed] [Google Scholar]
- 97. Izquierdo JM. Hu antigen R (HuR) functions as an alternative pre-mRNA splicing regulator of Fas apoptosis-promoting receptor on exon definition. J Biol Chem 2008;283:19077–84. [DOI] [PubMed] [Google Scholar]
- 98. Chang SH, Elemento O, Zhang J, Zhuang ZW, Simons M, Hla T. ELAVL1 regulates alternative splicing of eIF4E transporter to promote postnatal angiogenesis. Proc Natl Acad Sci U S A 2014;111:18309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wang Y, Guo Y, Tang C, Han X, Xu M, Sun J, et al. Developmental cytoplasmic-to-nuclear translocation of RNA-binding protein HuR is required for adult neurogenesis. Cell Rep 2019;29:3101–17. [DOI] [PubMed] [Google Scholar]
- 100. Fu XD, Ares M Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 2014;15:689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Saez B, Walter MJ, Graubert TA. Splicing factor gene mutations in hematologic malignancies. Blood 2017;129:1260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wan Y, Wu CJ. SF3B1 mutations in chronic lymphocytic leukemia. Blood 2013;121:4627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Papaemmanuil E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med 2016;374:2209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Saygin C, Hirsch C, Przychodzen B, Sekeres MA, Hamilton BK, Kalaycio M, et al. Mutations in DNMT3A, U2AF1, and EZH2 identify intermediate-risk acute myeloid leukemia patients with poor outcome after CR1. Blood Cancer J 2018;8:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ghigna C, Moroni M, Porta C, Riva S, Biamonti G. Altered expression of heterogenous nuclear ribonucleoproteins and SR factors in human colon adenocarcinomas. Cancer Res 1998;58:5818–24. [PubMed] [Google Scholar]
- 106. Stickeler E, Kittrell F, Medina D, Berget SM. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene 1999;18:3574–82. [DOI] [PubMed] [Google Scholar]
- 107. He X, Ee PL, Coon JS, Beck WT. Alternative splicing of the multidrug resistance protein 1/ATP binding cassette transporter subfamily gene in ovarian cancer creates functional splice variants and is associated with increased expression of the splicing factors PTB and SRp20. Clin Cancer Res 2004;10:4652–60. [DOI] [PubMed] [Google Scholar]
- 108. Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat Med 2017;23:1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Jones CL, Stevens BM, D'Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell 2018;34:724–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem 2017;86:685–714. [DOI] [PubMed] [Google Scholar]
- 111. Hope KJ, Cellot S, Ting SB, MacRae T, Mayotte N, Iscove NN, et al. An RNAi screen identifies Msi2 and Prox1 as having opposite roles in the regulation of hematopoietic stem cell activity. Cell Stem Cell 2010;7:101–13. [DOI] [PubMed] [Google Scholar]
- 112. Picelli S, Faridani OR, Bjorklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 2014;9:171–81. [DOI] [PubMed] [Google Scholar]
- 113. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7:562–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 shows expression of RBPs in an RNA-sequencing data set of human bone marrow hematopoietic stem and progenitor subpopulations.
Table S2 shows sequences of (a) sgRNAs from the RBP screen, (b) sequencing primers, (c) shRNAs and human-targeting sgRNAs, (d) eCLIP oligos.
Table S3 shows clinical information for all AML patient specimens used in this study.
Table S4 shows limiting dilution analysis of secondary recipients transplanted with DMSO- or MS-444-treated bone marrow from primary mice
Table S5 shows the log2 fold change of differentially expressed transcripts from the ELAVL1 knockout RN2c RNA-sequencing data set.
Table S6 shows altered gene sets identified by GSEA of the ELAVL1 knockout RN2c RNA-sequencing data set.
Table S7 shows (a) reproducible peaks and (b) GO analysis of ELAVL1-bound transcripts identified in the eCLIP-sequencing data set. The negatively enriched (c) and positively-enriched (d) gene sets identified by GSEA of ELAVL1-bound and differentially expressed transcripts from the RN2c RNA-sequencing data set are shown.
Table S8 shows transcripts from the ELAVL1 knockout RNA-sequencing data set that are (a) alternatively spliced and (b) alternatively spliced as well as bound by ELAVL1 as identified by eCLIP-sequencing.
Table S9 shows the (a) differentially expressed transcripts and (b) altered pathways as identified by GSEA from the ELAVL1 knockdown human primary AML RNA-sequencing data set.
Supplemental Figure S1 shows expression of RBPs in healthy HSPCs and RN2c cells, sgRNA drop out dynamics in primary and secondary transplants, the molecular functions and biological processes associated with the hit RBPs and their gene essentialities.
Supplemental Figure S2 shows validation of sgRNA -mediated knockout of hit RBPs, comparison of hit RBP depletion in RN2c in vitro versus in vivo, effects of ELAVL1 loss on human AML cell lines, characteristics of the MLL-AF9 AML mouse model, and ELAVL1 expression in normal hematopoietic cells.
Supplemental Figure S3 shows the generation of the bcCML mouse model and the effects of ELAVL1 on normal bone marrow in vivo.
Supplemental Figure S4 shows the validation of ELAVL1 knockdown by western blot and effects of ELAVL1 loss on human primary AML cells in vitro and in vivo.
Supplemental Figure S5 shows the effects of DHTS treatment on THP-1 and human primary AML cells in vitro, the effects of MS-444 treatment on human umbilical cord blood in vivo, and validation of ELAVL1 overexpression by western blot.
Supplemental Figure S6 shows the GSEA of ELAVL1 depletion in mouse RN2c and human primary AML cells, ELAVL1 subcellular localization in mouse and human AML, the qRT-PCR of ELAVL1 targets in MS-444-treated RN2c cells, and the integrative analysis of the mouse RNA-seq and eCLIP-seq.
Supplemental Figure S7 shows the expression profile of TOMM34 in normal human bone marrow and across different AML and CML data sets. The effects of TOMM34 depletion on human primary AML cells in vitro and validation of TOMM34 knockdown and overexpression is shown.
Data Availability Statement
The eCLIP and mouse RNA-seq data generated in this study were deposited and are publicly available in Gene-Expression Omnibus (GEO) at accession references GSE127944 and GSE127743, respectively. Human AML RNA-seq data set: GSE224548. All other data supporting the findings of this study are cited in Materials and Methods, in supplementary documents, or are available upon request from the authors.