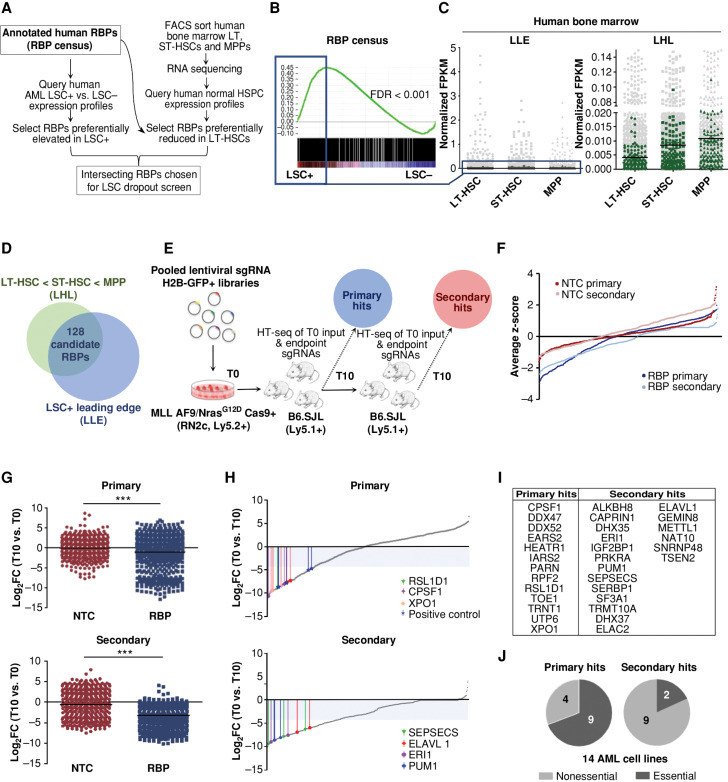
Figure 1.
Diverse RBPs are enriched in LSCs and identified as in vivo AML LSC essentialities through a two-step pooled in vivo CRISPR-Cas9 dropout screen. A, Overview of in silico selection of RBPs preferentially heightened in LSCs and reduced in LT-HSCs. B, GSEA plot showing LSC-enriched RBPs. The top 500 of the leading-edge LSC-enriched RBPs (LLE) are indicated by the blue box. C, Expression of LSC-enriched RBPs (LLE) in LT-HSC, ST-HSC, and MPP populations of human BM (dark gray, left) relative to all RBPs in census (light gray, left) and a subset of RBPs (LHL) with lowest expression in LT-HSCs of human BM (green, right) relative to all RBPs in census (light gray, right). D, Selection of the 128 RBPs exhibiting LSC-enriched and LT-HSC-reduced expression. E, Schematic illustrating the in vivo dropout screen. HT-seq, high-throughput sequencing. F, Average ranked dropout z-scores for all sgRNAs in both arms and transplantation rounds. G, Median log2 fold-change (T10 vs. T0) of unique sgRNAs in the NTC and RBP arms of the screen at the primary (top) and secondary (bottom) endpoints are shown. H, Median LFC of all sgRNAs within the RBP arm of the screen after the primary (top) and secondary (bottom) rounds. Select top-scoring sgRNAs are indicated with colored bars; shaded area indicates the decreased fold change of 20 cutoff. I, RBPs are called hits across primary and secondary screening arms. J, Analysis of general essentiality across a panel of AML cell lines (40) for the RBPs considered hits at primary endpoints (primary, left) compared with RBP hits where all targeting sgRNAs dropped out only in secondary recipients (secondary, right). A gene was considered generally essential if its average log2 fold change in abundance was less than −1 in at least 12 of the 14 tested AML lines in vitro. n = 2–3 mice per 1° T10 and 2° T0 and T10 replicates. LT-HSC = CD34+CD38−CD90+CD49f+; ST-HSC = CD34+CD38−CD90+CD49f−; MPP = CD34+CD38−CD90−CD49f−. ***, P < 0.001, determined by a two-sided Student t test.