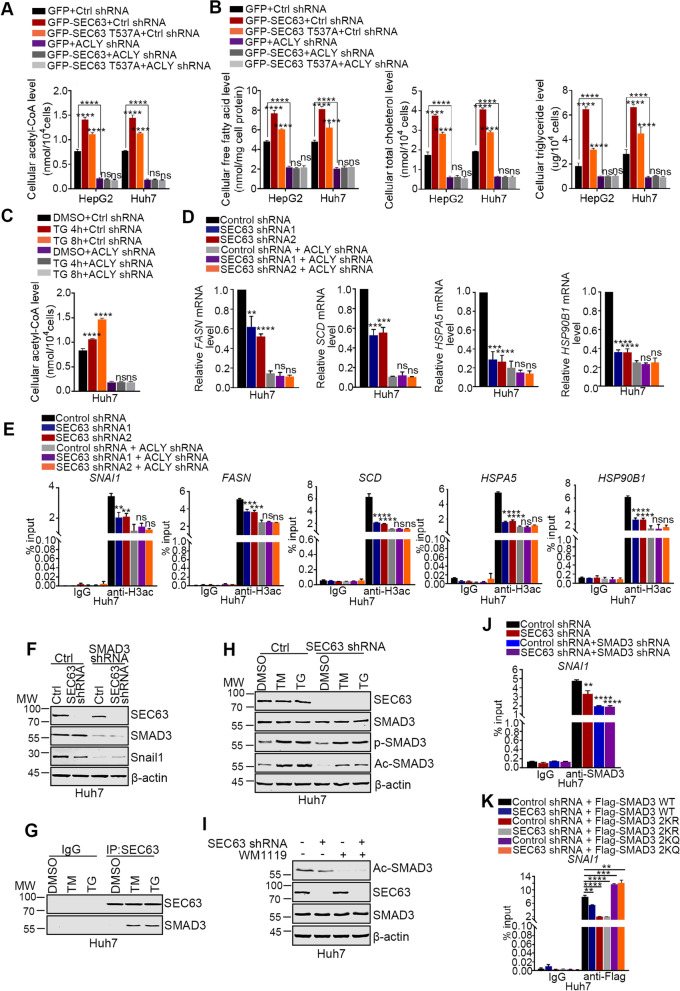
Fig. 7.
SEC63 modulated ACLY-related metabolic and epigenetic reprogramming. A The cellular acetyl-CoA level was evaluated in the HepG2 and Huh7 cells (n = 4). B The cellular free fatty acids, cholesterol, and triglyceride levels were evaluated in the HepG2 and Huh7 cells (n = 4). C The cellular acetyl-CoA level was detected in the Huh7 cells transfected as indicated (n = 4). D Total RNAs were extracted from the Huh7-ctrl or Huh7-shSEC63 cells transfected with ACLY shRNA. RT-qPCR was performed for the UPR target genes (FASN, SCD, HSPA5, HSP90B1) (n = 3). E ChIP-qPCR was performed using anti-acetyl-H3 antibodies (n = 3). F Western blot analysis of indicated protein levels in Huh7 cells. G The cells were treated with TM (5 μg/mL) or TG (1 μM) for 8 h and immunoprecipitation was performed with anti-SEC63 antibody. H The Huh7-ctrl or Huh7-shSEC63 cells were treated with TM or TG. Western blot was further performed using the indicated antibodies. I The ctrl cells or SEC63-depleted cells were treated with WM1119 (KAT6A inhibitor). Western blot was performed for the indicated proteins. J, K ChIP assays were performed with IgG, anti-SMAD3 or anti-Flag antibody followed by qPCR to amplify the SNAI1 promoter region. The data are shown as the mean ± SD