Fig. 2 |. Characterization of tissue-resident CD45+ cell populations in Efat by scrNA-seq.
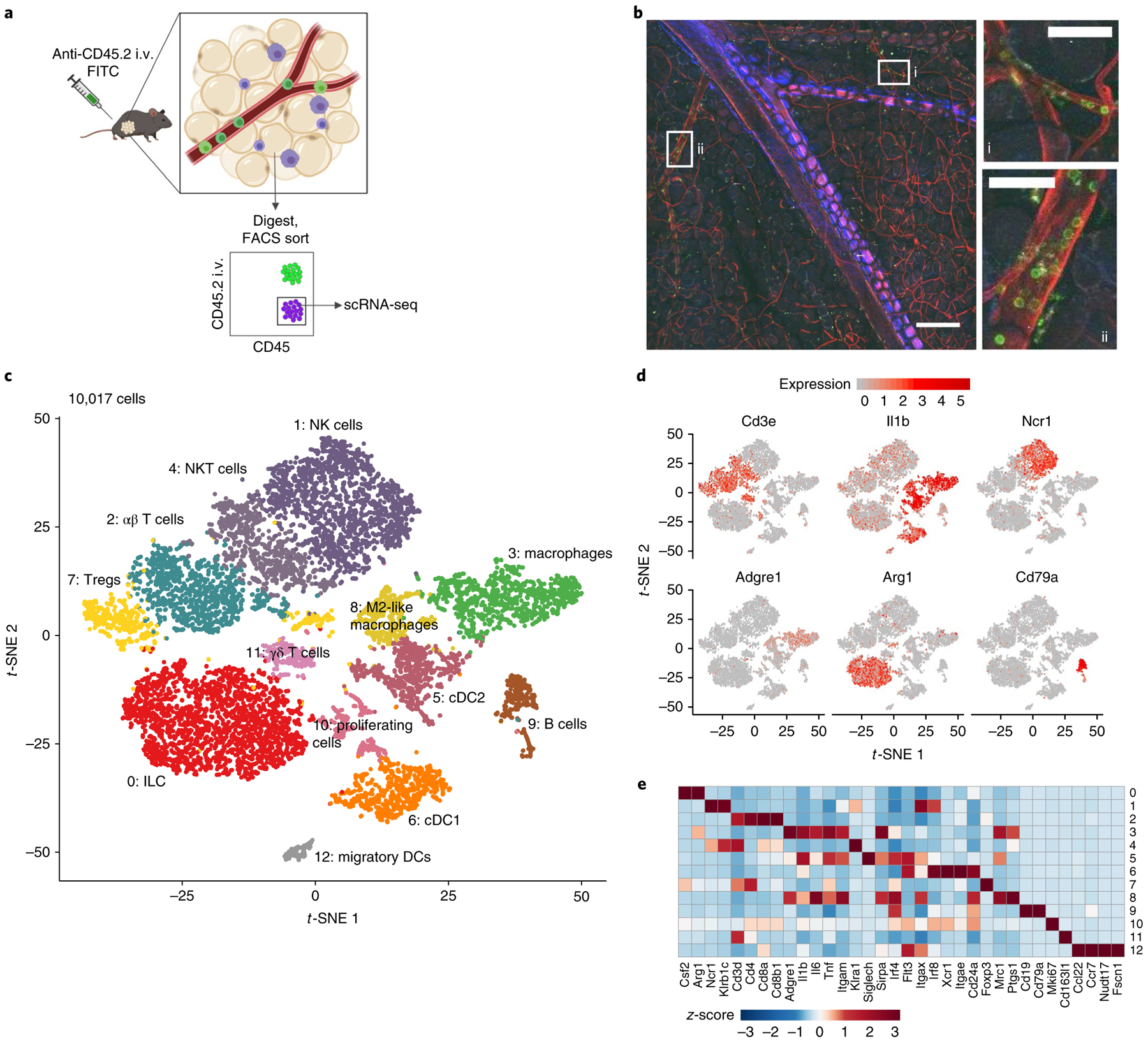
a, Schematic depicting cell sorting strategy to isolate tissue-resident haematopoietic cells in visceral adipose tissue. b, Two-photon microscopy showing i.v.-labelled CD45+ cells in Efat, scale bar, 200 μm. Insets highlight (i) microvasculature; scale bar, 50 μm and (ii) larger blood vessels within visceral adipose tissue; scale bar, 50 μm. Vasculature is visualized in red using a global membrane-Tomato reporter mouse, i.v.+ (circulating, non-resident) cells are shown in green after i.v. injection of anti-CD45.2-A488. Images are representative of two independent experiments. c, t-SNE plot of tissue-resident (i.v.−) CD45+ cells from Efat. Clustering is based on merged analysis of chow- and KD-fed mice. d, t-SNE plot as in c, displaying expression of selected lineage marker genes. e, Heat map of normalized gene expression values of selected genes to identify major lineages. For c–e, expression values were obtained by pooling data from chow and KD samples (each containing n = 4 chow and n = 3 KD pooled biological samples into one technical sample for each diet).
