Figure 1.
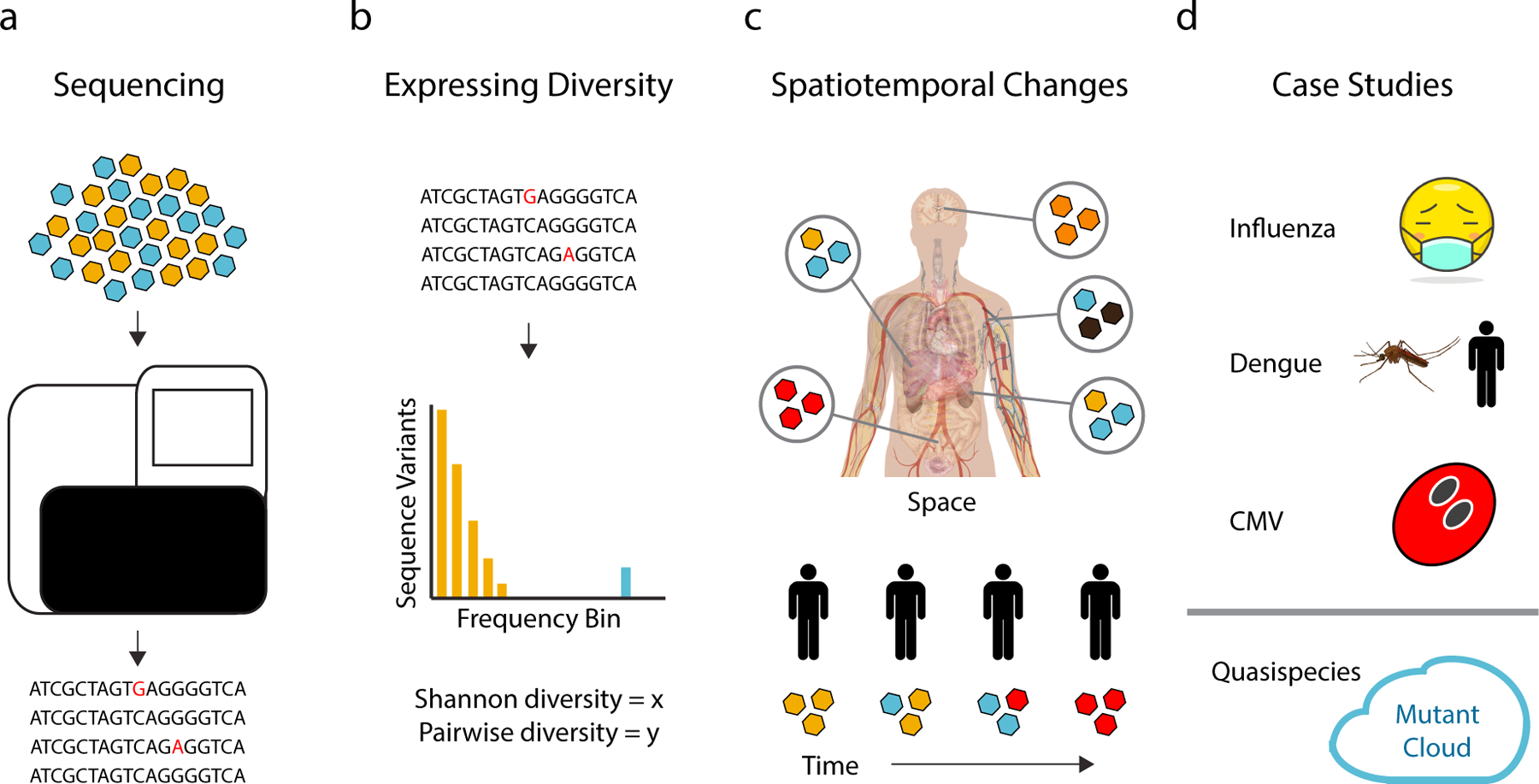
Within-host diversity and virus evolution. (a) Advances in sequencing technology have revolutionized the study of within-host viral diversity. The type and frequency of mutations (red letters) in a population are easily obtained from NGS (b) Viral diversity can be measured and compared using a variety of metrics. Ideally, these metrics capture the varying impact of mutations present at high (blue bars) vs. low (yellow bars) frequency. (c) Differences in diversity across tissues (top, different colored viruses) or changes over time (bottom) can be used to model within-host viral dynamics due to selection and genetic drift. (d) Studies of within-host viral diversity have provided insights into the evolution of influenza virus, Dengue virus, and cytomegalovirus and the extent to which within-host virus populations can be accurately described as “quasispecies” or “mutant clouds.”
