Figure 3.
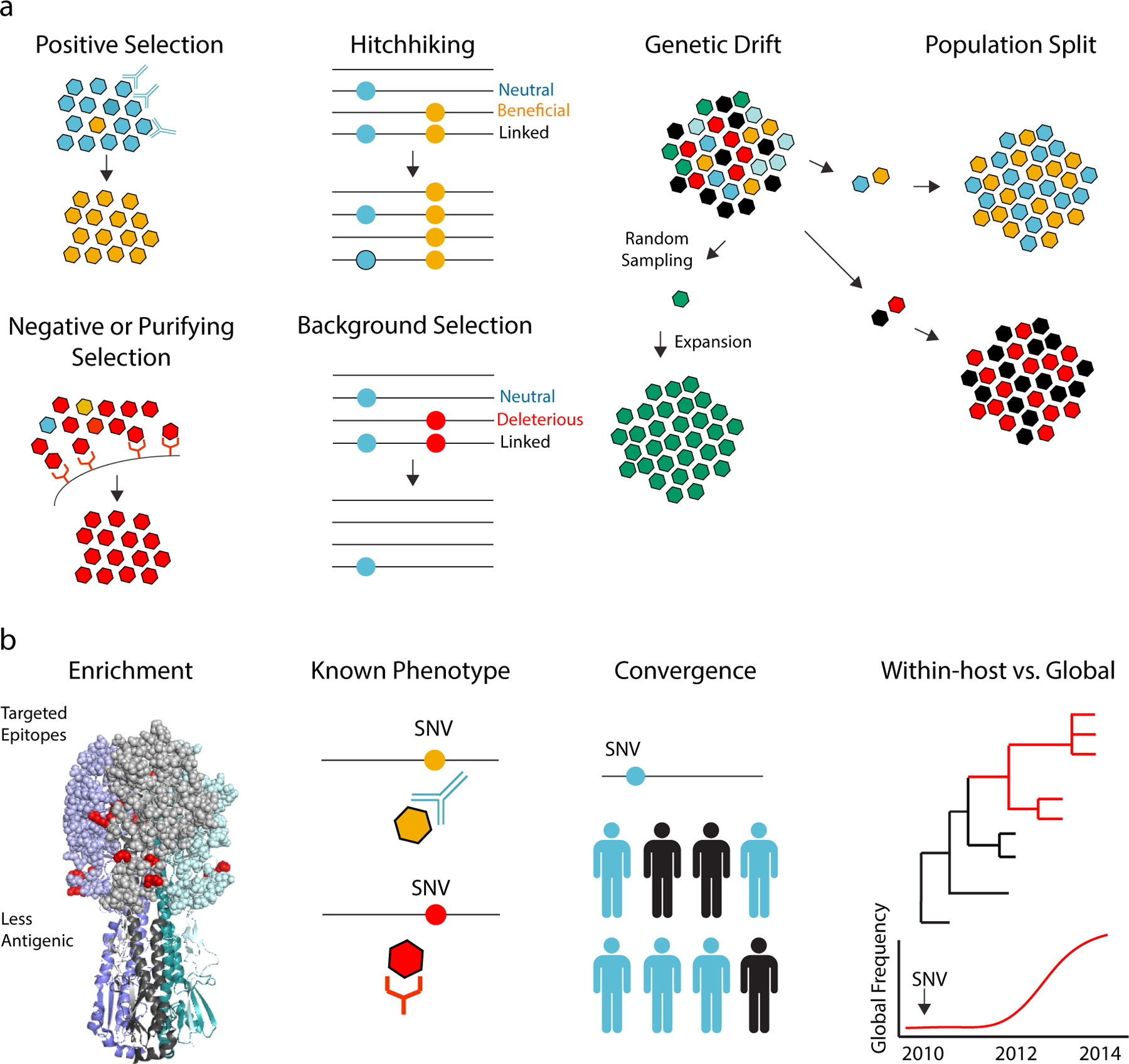
Using within-host data to elucidate evolutionary dynamics. (a) The frequency of a mutation can increase or decrease due to selective and nonselective processes. Positive selection will increase the frequency of a beneficial mutation (e.g., a mutation that leads to antibody escape) and will decrease the frequency of a detrimental mutation (e.g., a surface protein mutant that can no longer bind its receptor). A neutral mutation (blue) can increase in frequency if it is linked to a beneficial mutation on the same genome (hitchhiking) or decrease in frequency if it is linked to a detrimental one (background selection). Genetic drift is a change in a mutation’s frequency due to stochastic processes. It typically occurs in small populations due to random sampling. Genetic drift also occurs during population bottlenecks or splits followed by expansions. (b) Criteria for identifying within-host variants potentially under positive selection. From left to right: enrichment of high frequency variants in viral proteins or certain protein domains, shown here as nonsynonymous single nucleotide variants (SNV) in antigenic sites (red) modeled on the structure of the influenza hemagglutinin trimer; identification of a variant already known to mediate a certain phenotype, such as antibody escape or receptor binding; identification of the same mutation in multiple individual hosts not linked by transmission; identification of a within-host variant that is subsequently seen in different host populations or at different evolutionary scales.
