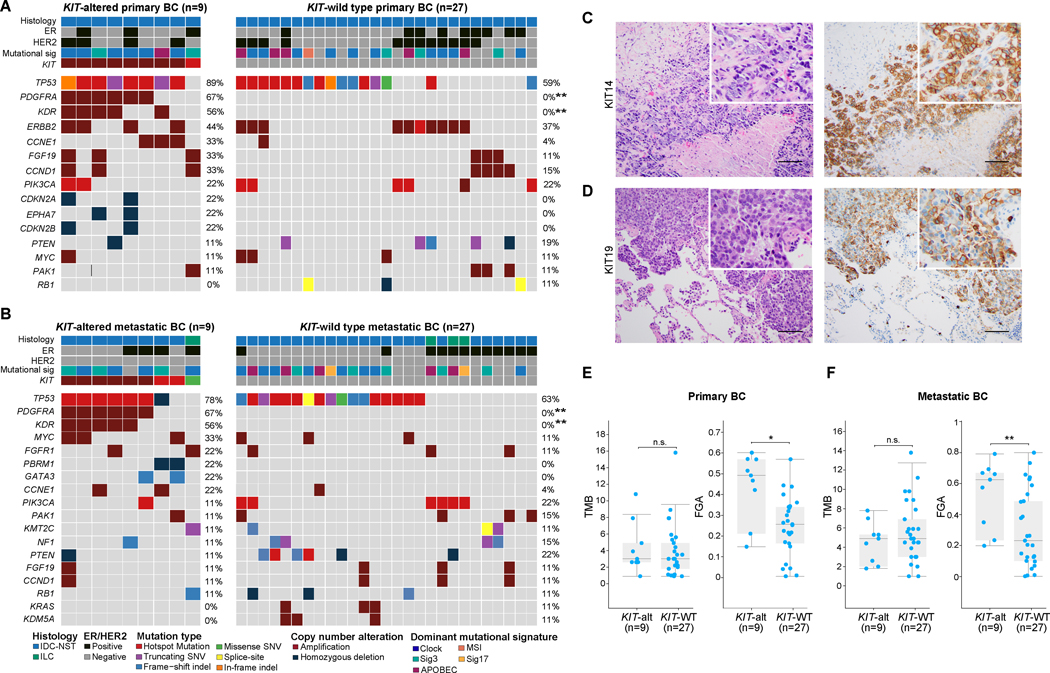
Figure 1. Repertoire of genetic alterations in breast cancers with KIT genetic alterations.
(A-B) Heatmaps depicting the non-synonymous somatic mutations, amplifications and homozygous deletions in (A) primary (n=9) and (B) metastatic (n=9) KIT-altered breast cancers (BC) compared to BCs lacking KIT genetic alterations matched by clinicopathologic features at a 3:1 ratio (n=27, primary and metastatic BC, each). Recurrently affected (≥2 cases) genes in KIT-altered BCs and the most frequently altered (≥3 cases) genes in KIT-wild type (WT) BCs are shown. Fisher’s exact test, FDR adjusted P value, **, <0.01. (C-D) Representative hematoxylin and eosin (H&E) micrographs (left) and corresponding KIT protein expression micrographs (right) assessed by immunohistochemistry for the (C) primary BC case KIT14, and (D) metastatic BC case KIT19. Scale bar, 50 microns. (E-F) Box-plots displaying the non- synonymous tumor mutation burden (TMB; left) and fraction of genome altered (FGA; right) in (E) primary and (F) metastatic BCs harboring KIT genetic alterations (n=9, each) compared to KIT-WT BCs matched by clinicopathologic features at a 3:1 ratio (n=27, each). Mann-Whitney U test, *, P<0.05; **, P<0.01; n.s., non-significant.