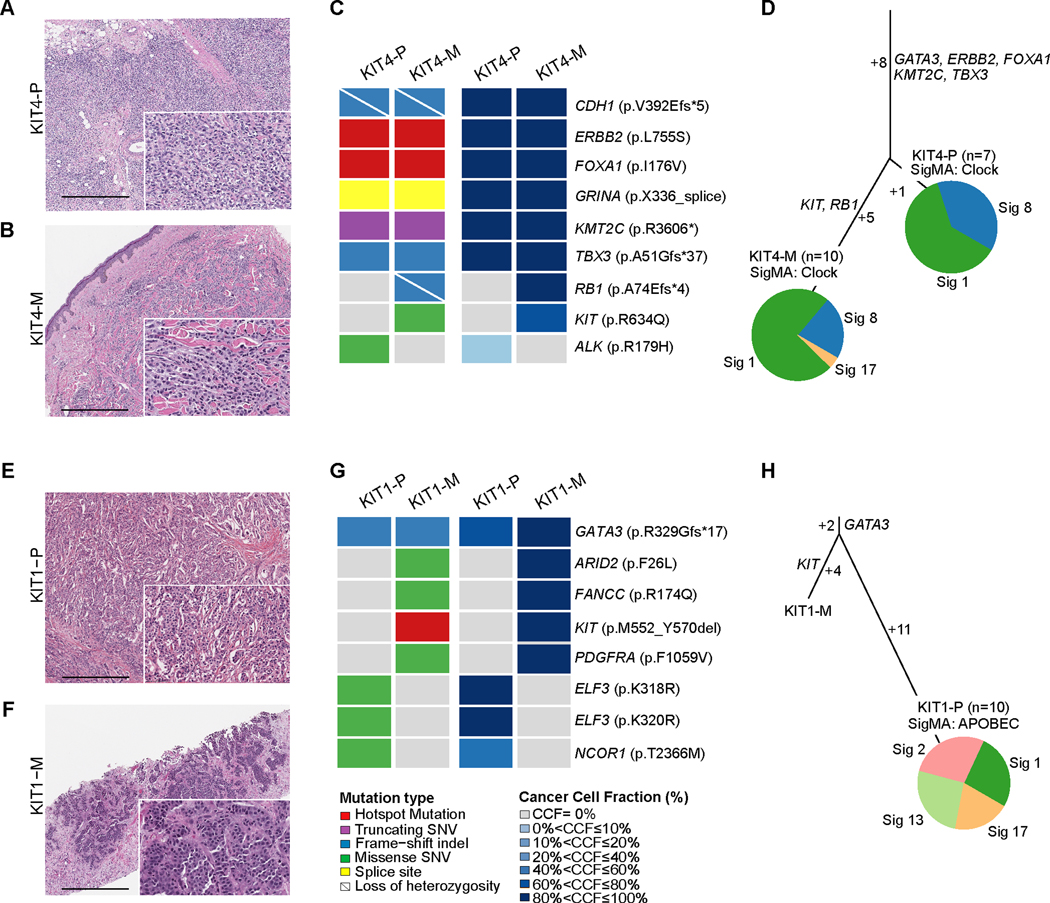
Figure 2. Clonal decomposition and mutational signatures of paired primary and metastatic KIT-altered breast cancers.
(A,B,E,F) Representative hematoxylin and eosin (H&E) micrographs of the paired primary (P) and metastatic (M) breast cancer (BC) samples of case KIT4 including (A) KIT4-P and (B) KIT4-M, and of case KIT1, including (E) KIT1-P and (F) KIT1-M. Scale bar, 500 microns. (C,G) Heatmaps depicting the non-synonymous somatic mutations (left) and cancer cell fraction (right) of the paired primary and metastatic BC samples of cases (C) KIT4 and (E) KIT1. (D,H) Mutation based phylogenetic trees depicting the clonal evolution of the paired primary and metastatic BC samples of (D) case KIT4 and (H) case KIT1. The length of the trunk and branches of the trees is proportional to the number of shared and private mutations in the primary and metastatic BC samples. Mutational signatures identified in the primary and metastatic BCs with ≥5 SNVs as inferred by SigMA are depicted in pie charts.