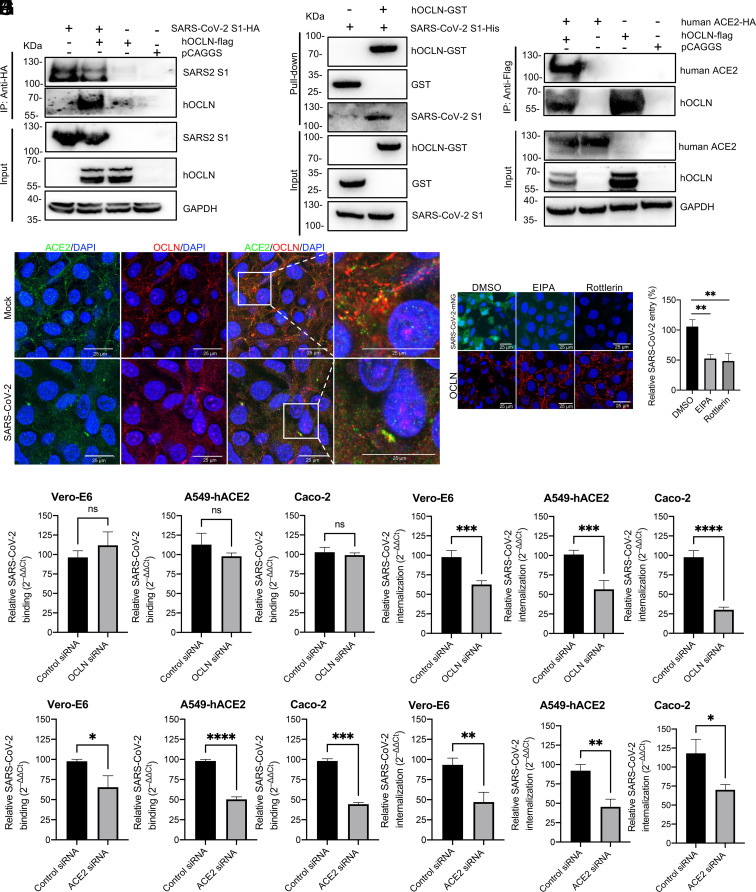
Fig. 3.
SARS-CoV-2 S protein directly interacts with OCLN and SARS-CoV-2 internalization but not virus binding requires OCLN. (A) SARS-CoV-2 S1 tagged with HA and human OCLN plasmids tagged with Flag were cotransfected into 293T cells. At 72 h posttransfection, the cell lysate was harvested. Co-IP was performed using an anti-HA antibody. The precipitated proteins were detected by western blot using antibodies against the HA and Flag tags. (B) For the GST pull-down assay, GST and GST-tagged hOCLN were expressed in Escherichia coli BL21(DE3) and purified with glutathione resin. The resin was incubated with purified SARS-CoV-2 His-S1 protein, and the bound proteins were detected by western blot using an anti-GST antibody and an anti-His antibody. (C) Interaction between OCLN and ACE2. Human ACE2 plasmid tagged with HA and human OCLN plasmid tagged with Flag were cotransfected into 293T cells. 72 h posttransfection, the cell lysate was harvested. Co-IP was performed by using anti-Flag antibody. The precipitated proteins were detected by western blot with antibodies against the HA and Flag tags. (D) Colocalization of OCLN and ACE2. Vero-E6 cells were mock-infected or infected with SARS-CoV-2 at an MOI of 0.1. 48 h postinfection, the cells were fixed. ACE2 and OCLN were detected by an anti-ACE2 and an anti-OCLN antibody. Images were acquired with a confocal microscope. (E) Virus binding assay: Vero-E6, A549-hACE2, or Caco-2 cells were infected with SARS-CoV-2 at an MOI of 1 at 4 °C for 1 h. Unbound viruses were removed by washing with cold phosphate buffered saline (PBS), and cells then treated with 1 mL TRIzol to extract RNA for RT-qPCR. (F) Virus internalization assay: Cells were transfected with OCLN or control siRNA and then infected with SARS-CoV-2 at an MOI of 1 at 4 °C for 1 h. Unbound viruses were removed with cold PBS. The cells were transferred to 37 °C for 1 h to allow virus internalization and then washed to remove bound virus on cell surface with acidic buffer. The cells were lysed, and RNA was prepared for RT-qPCR. ACE2 knockdown with specific siRNA was used as a positive control for virus binding assay (G), and virus internalization assay (H) was performed. (I) Vero-E6 cells were treated with micropinocytosis inhibitors, EIPA, or rottlerin for 1 h prior to SARS-CoV-2-mNG infection. Forty-eight hours postinfection, virus infection was observed through GFP signals, and OCLN distribution was detected with IFA by using an anti-OCLN antibody. (J) Vero-E6 cells were pretreated with EIPA or rottlerin and then infected with SARS-CoV-2 at 4 °C for 1 h. Unbound viruses were removed with the cold PBS. The cells were transferred to 37 °C for 1 h to allow virus internalization and then washed to remove bound virus on cell surface with acidic buffer. The cells were lysed, and RNA was prepared for RT-qPCR to determine virus entry.