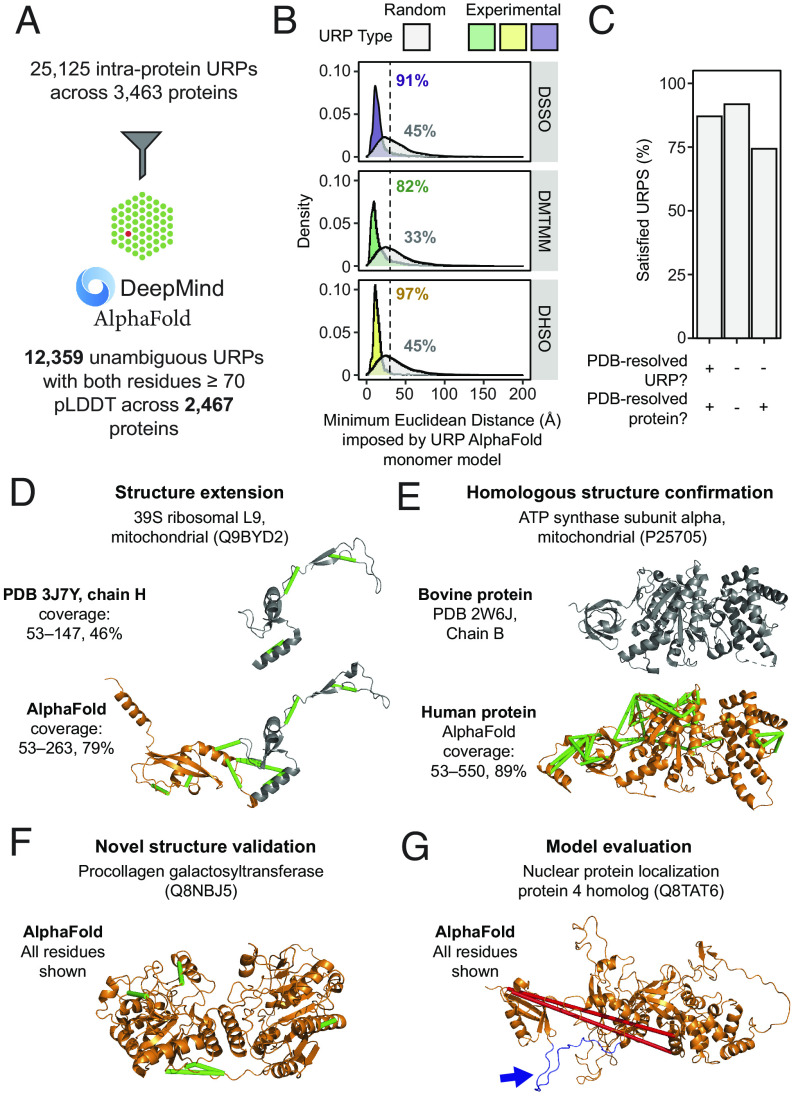
Fig. 3.
The high-throughput experimental assessment of AlphaFold2 monomeric protein structure predictions using cross-link distance constraints. (A) Statistics for the mapping of unique URPs onto AlphaFold2 (AF2) monomer models. Only high-confidence URPs (both residues within pLDDT ≥70) were used for further analyses. (B) Fulfillment of URPs on AF2 models. For each AF2 monomer model, five random URPs were generated for each cross-linker with appropriate side-chain reactivities. The distribution of Euclidean distances (Å, Cα-Cα) determined for experimental and random URPs is shown, stratified by cross-linker. Annotations show the percentage of URPs fulfilled for each subset. The cutoff for each cross-linker is indicated by a dotted line (25 Å for DMTMM or 30 Å for DHSO/DSSO). (C) Overall URP satisfaction rate across URPs with different degrees of PDB resolution. (D–G) Examples of the use of URPs for model validation and evaluation. Protein names are official UniProt entry names. The range of residues shown are indicated to the left of each structure. Green URPs are satisfied, red URPs are overlength. (D) The model for Q9BYD2 shows an example of a PDB-resolved human protein that has benefitted from increased structural coverage (orange) and is now corroborated by nine experimental URPs. Gray regions within structures represent those derived from experimental PDB structures. (E and F) show proteins without any PDB entry for the human proteins (orange). Gray regions within structures represent those derived from experimental PDB structures. (E) The AF2 model of the mitochondrial ATP synthase subunit alpha with a known bovine structural homologue, 44 URPs support this model. (F) The AF2 model of the procollagen galactosyltransferase (Q8NBJ5) that does not have any PDB entries for itself or of homologous proteins. Five URPs support this model. (G) An example where the two identified cross-links do not fit the AF2 model. However, while the cross-linked residues are within well-modeled domains, the two domains are separated by a low-confidence, disordered loop (colored in blue and highlighted with a blue arrow).