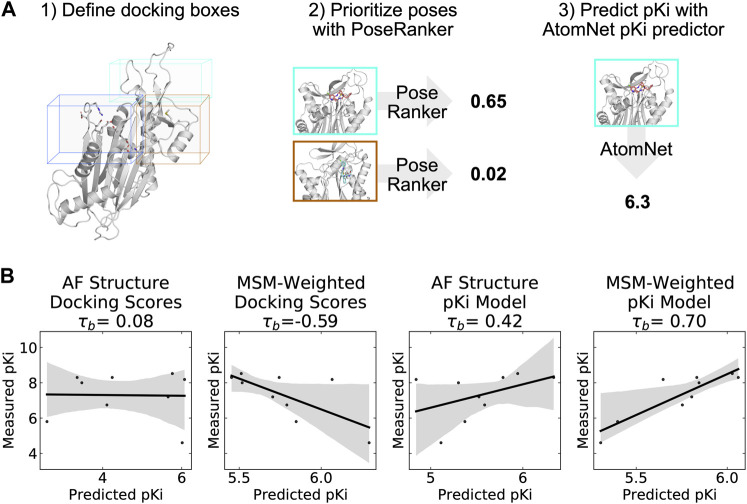
FIGURE 5.
A neural network trained to predict pKi accurately ranks allosteric compounds by potency when applied to structures from a PPM1D ensemble. (A) Schematic highlighting the procedure that was used for selecting a single pose for each PPM1D cluster center in the MSM. For each MSM cluster center, we defined multiple docking boxes based on the active site, residues involved in photolabeling experiments, and P2Rank pockets at the flap and hinge. After performing docking, we selected a best pose per MSM state using the PoseRanker neural network. Finally, we fed this best docked pose to the AtomNet pKi predictor. (B) MSM-weighting of the pKi predictions from the AtomNet pKi predictor outperforms docking-based methods as well as a single pKi prediction based on the AlphaFold-predicted structure. For each scatter plot, we show the line of best fit in black as well as the 95% confidence interval based on bootstrapping in translucent grey bands. We report the Kendall rank correlation coefficient, a statistic that measures the ordinal association between the predited pKi and the measured pKi and whose maximum value is 1.