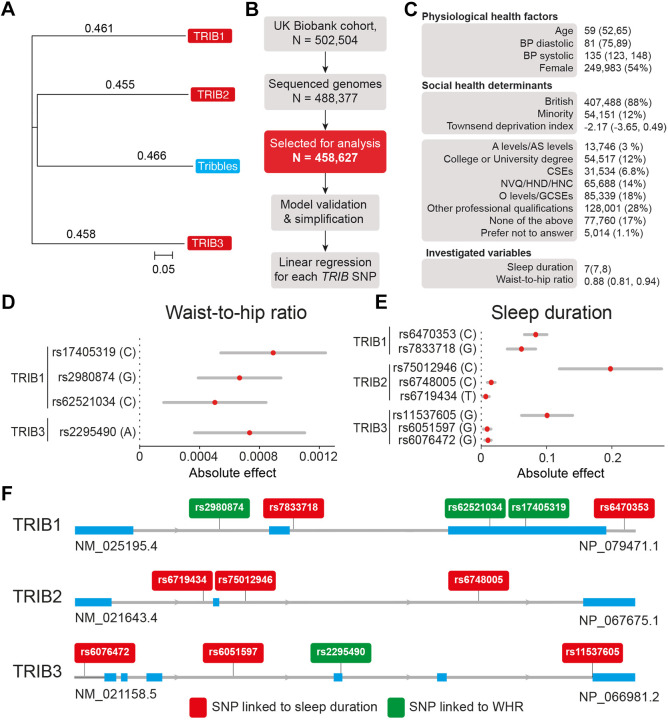
Fig. 5.
Polymorphisms in human tribbles pseudokinase genes (TRIB1-3) are associated with alterations in sleep and obesity in the UK Biobank cohort. (A) The Drosophila tribbles gene shows highest conservation with human TRIB2. Phylogenetic analysis between Drosophila tribbles (blue) and human TRIB1-3 (red) was performed using the Neighbour Joining method. The scale corresponds to the distance between proteins in phylogenetic units, where a value of 0.05 corresponds to a difference of 5% between two sequences. (B) Workflow for the analysis of the participants of the UK Biobank cohort. After excluding participants with incomplete data, a subset of 458,627 individuals containing genomic and phenotypic data were selected for further analysis. (C) Descriptive statistics of the participants analysed in this study. Either the number of participants in each category and their percentage with respect to the total cohort, or the median and interquartile ranges are shown. (D,E) Absolute effect size of the significant SNPs in human TRIB genes associated with obesity (D) or sleep (E), following FDR correction using the Benjamini–Hochberg method. The red circles correspond to the absolute effect size of the SNP and the grey bars are 95% confidence intervals. (F) Genomic location of the TRIB SNPs that are significantly associated with sleep (red) and obesity (green). Blue corresponds to exons and grey to intronic regions.