Figure 1. The pipeline of the APEAL approach.
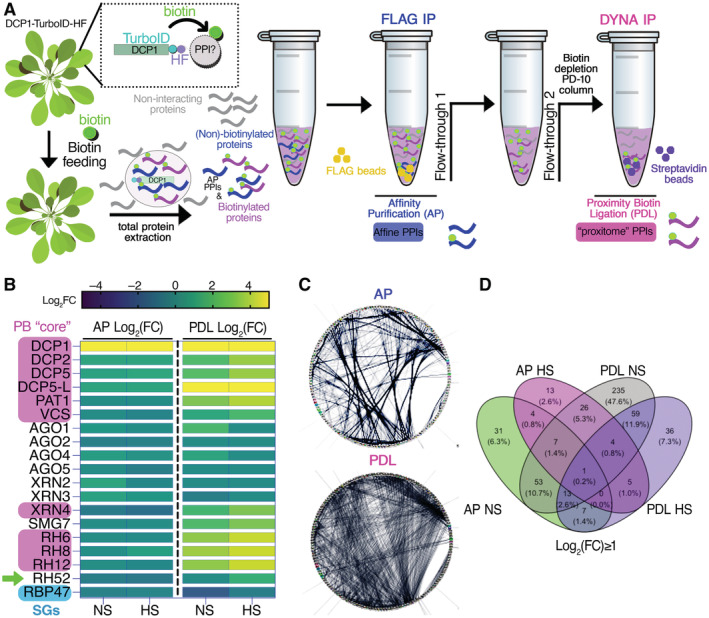
- Overview of the APEAL pipeline. Upon 24 h biotin feeding and treatment (supplied with 50 μM biotin directly into leaves of 4‐week‐old plants by syringe infiltration), total proteins are extracted from infiltrated leaves. The proteome is subjected to AP‐immunocapture of the FLAG tag. In the AP step, some of the captured proteins will be biotinylated. The PDL step uses the leftover supernatant from the AP step and captures biotinylated proteins with streptavidin beads. PPI, protein–protein interactions; AP, affinity purification; PDL, proximity‐dependent biotin ligation; FLAG IP, FLAG‐beads immunoprecipitation; DYNA‐IP, streptavidin‐bead immunoprecipitation. We use the term “proxitome,” to describe the proteins captured by the PDL step of APEAL. These proteins may not physically interact with DCP1.
- Heatmap showing the “core processing body (PB)” (magenta) components identified and other linked proteins. RBP47 is a stress granule marker (SGs; blue). RH52 is a new helicase identified as a PB component (green arrow). The scale on the right shows log2FC of protein abundance. Note that only the PB core components were enriched in the PDL (i.e., the proxitome, log2FC ~ 1 or above) but not in AP. Furthermore, heat stress (HS) increased the enrichment of some PB core components.
- Comparison of AP/PDL interacting networks produced from APEAL. STRING density plots of pairwise interactions between proteins obtained from the AP or PDL steps (combined interactions found in non‐stress [NS]/[HS]). Note that PDL produces an overall denser interaction network (under standard parameters, the same number of proteins was selected for AP/PDL).
- Venn diagram showing the proteins identified for PDL and AP in NS and HS samples (PPIs fulfilling the criterion log2FC > 1).
Source data are available online for this figure.
