Figure EV3. Hits from the APEAL approach (AP and PDL steps).
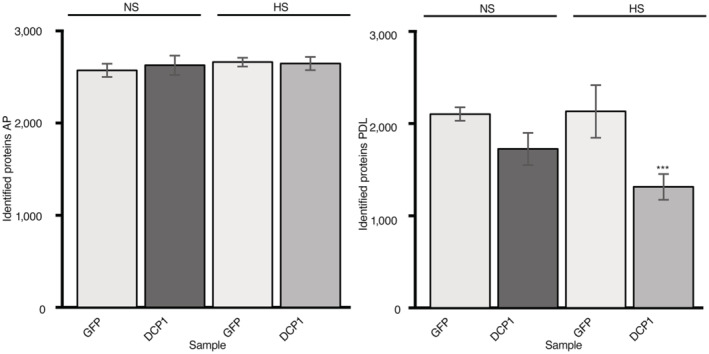
Total protein hits from mass spectrometry under NS or HS conditions following the AP (left) or PDL (right) steps of APEAL. We used the same scheme for biotin application, as described (Arora et al, 2020). The results presented are unfiltered, containing the noisy portion of the proteome. Note that the free diffusion of GFP in vivo led to increased proteins identified. sGFP‐TurboID‐HF, GFP; DCP1‐TurboID‐HF, DCP1 (N, biological replicates = 3, error bars are mean ± SD). Note the increased numbers of hits in GFP/PDL reflect the noisy proteome. GFP is expected to produce more noise (translated as hits in the context of the proteome), due to the increased diffusion over the specifically and topologically restricted DCP1 (confirmed in Fig EV1, localizations). The decreased number of hits for HS conditions corresponds mainly to proteins of signal transduction and metabolism, as well as vesicle trafficking proteins (see also Fig 2 and below for an explanation: HS reduces the DCP1 association with the PM). Furthermore, DCP1, as described below, loses localization at the PM during HS.
Data information: AP/PDL GFP samples did not differ at P < 0.005 (determined by an unpaired t‐test); ***P ≤ 0.05, as determined by an unpaired t‐test for comparison between HS and NS GFP samples.
