Figure EV2. Bacterial aggregation enhances the uptake‐dependent killing of macrophages and bacteria propagation.
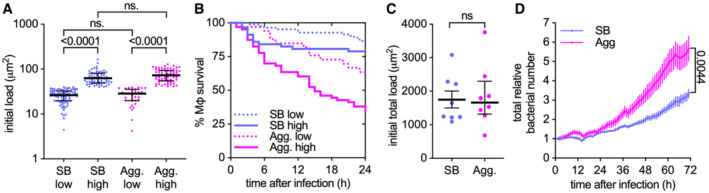
- Infected individual macrophages are binned into low and high initial loads according to the amount of single (SB) or aggregated (Agg) bacteria they internalize. The bacterial load is calculated as fluorescent area per macrophage and gates are set at < 40 μm2 (low) or > 40.0 μm2 (high) per macrophage. The area of one bacterium is included between 0.5 and 2 μm2. Each symbol represents the bacterial load of one individual macrophage (n = 82, 57, 33, and 63 macrophages, respectively). Bars represent the median and interquartile range. P‐values were calculated using a Krustal–Wallis test; ns, P‐values > 0.05.
- Percentage survival over time for individual macrophages with an initial bacterial load as indicated in panel A (n = 82, 57, 33, and 63 macrophages, respectively).
- Total initial bacterial load per microscopy field of view (332.80 × 332.80 μm2, approx. 100 cells/field of view). Each symbol represents one field of view (n = 8). Bars represent the mean and standard errors of the mean. P‐value calculated using an unpaired t‐test; P‐values > 0.05.
- Total relative bacterial load over time per microscopy field of view. Symbols represent the average bacterial load (n = 8) and bars represent standard errors of the mean. P‐values were calculated using an unpaired t‐test.
Source data are available online for this figure.
