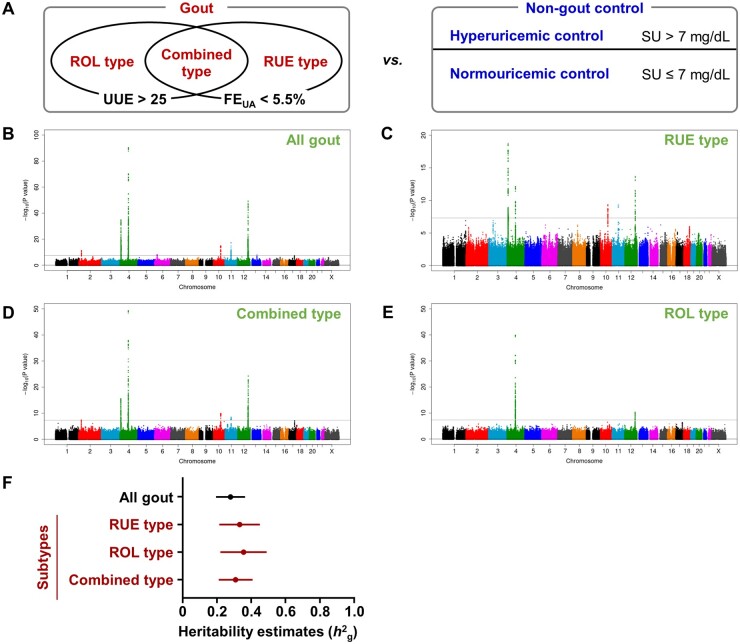
Figure 1.
Overview of the results of meta-analyses and SNP-based heritability estimates of gout. (A) Design of GWASs (gout cases vs non-gout controls) conducted in this study. (Left) Subtype classification of gout. UUE, urinary urate excretion (mg/h/1.73 m2); FEUA, fractional excretion of uric acid (%). FEUA was calculated using the following equation: ([urine urate]/[serum urate]) × ([serum creatinine]/[urine creatinine]) × 100. (Right) Classification of non-gout controls. In addition to normouricaemic subjects without gout characterized by serum urate (SU) ≤7 mg/dL (normouricaemic control), hyperuricaemic subjects without gout characterized by SU >7 mg/dL (hyperuricaemic control) were employed. (B–E) Manhattan plots of genome-wide meta-analyses for genetic loci that influence the risk of gout: (B) all gout, (C) RUE type, (D) combined (RUE + ROL) type and (E) ROL type. SNPs with a minor allele frequency ≥1% were analysed. The horizontal axis represents chromosomal positions; the vertical axis indicates the −log10(P-value) for assessment of the association. Horizontal lines represent the genome-wide significance threshold (α = 5 × 10−8). Information on significant loci identified in the present genome-wide meta-analysis is summarized in Supplementary Table S5, available at Rheumatology online. (F) Estimation of the SNP-based heritability of gout and its subtypes using clinically defined gout patients in Japanese populations (vs non-gout controls). Horizontal bars indicate the standard error of heritability. Further information, including the prevalence of gout and each subtype, and a comparison with the SNP-based heritability estimates of gout (vs normouricaemic controls) are shown in Supplementary Table S1, available at Rheumatology online