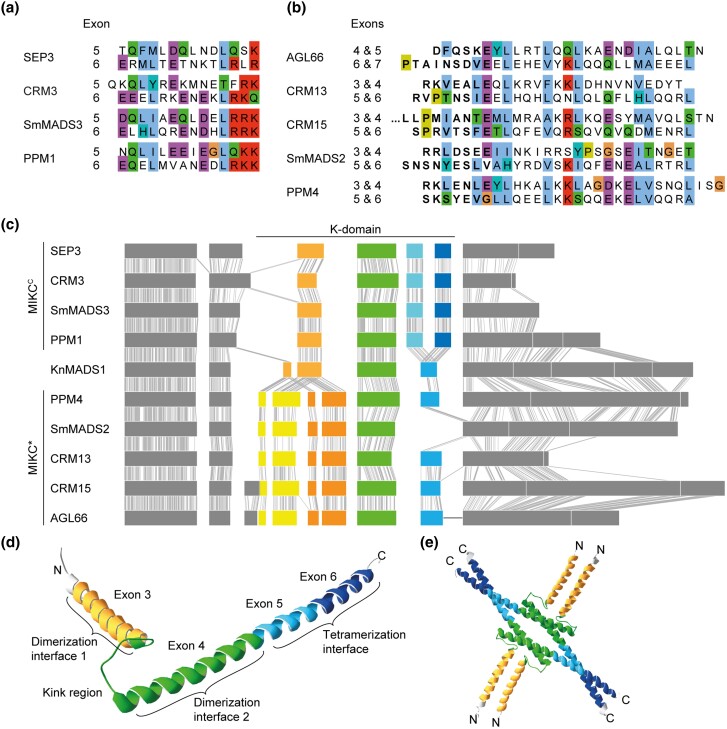
Fig. 3.
Similarity of K-domain exons hypothesized to be duplicated. (a, b) Multiple sequence alignment of the amino acids encoded by (a) exons 5 and 6 of the MIKCC-type genes SEP3, CRM3, SmMADS3, and PPM1 and (b) exons 4–7 of the MIKC*-type gene AGL66 and exons 3–6 of CRM13, CRM15, SmMADS2, and PPM4, respectively. (c) Exon–intron structure of the MIKCC- and MIKC*-type genes shown in a and b, respectively, together with the exon–intron structure of the charophyte MIKC-type gene KnMADS1. Homologous exons were aligned and identical nucleotide positions of neighboring sequences are connected with solid gray lines to illustrate homology. K-domain exons shared by MIKCC-, MIKC*-, and charophyte MIKC-type genes are color-coded in green, hypothetically duplicated K-domain exons of MIKCC- and MIKC*-type genes are color-coded in different shades of blue and yellow/orange, respectively. (d) X-ray crystal structure of the MIKCC-type protein SEP3 (PDB-ID: 4OX0). Subdomains encoded by exons 3, 4, 5, and 6, as indicated, are color-coded in yellow, green, light blue, and dark blue, respectively. (e) Tetramer of four SEP3 K-domains following the same color-coding as in d. Protein structure images were generated with Swiss-PdbViewer (Guex and Peitsch 1997).