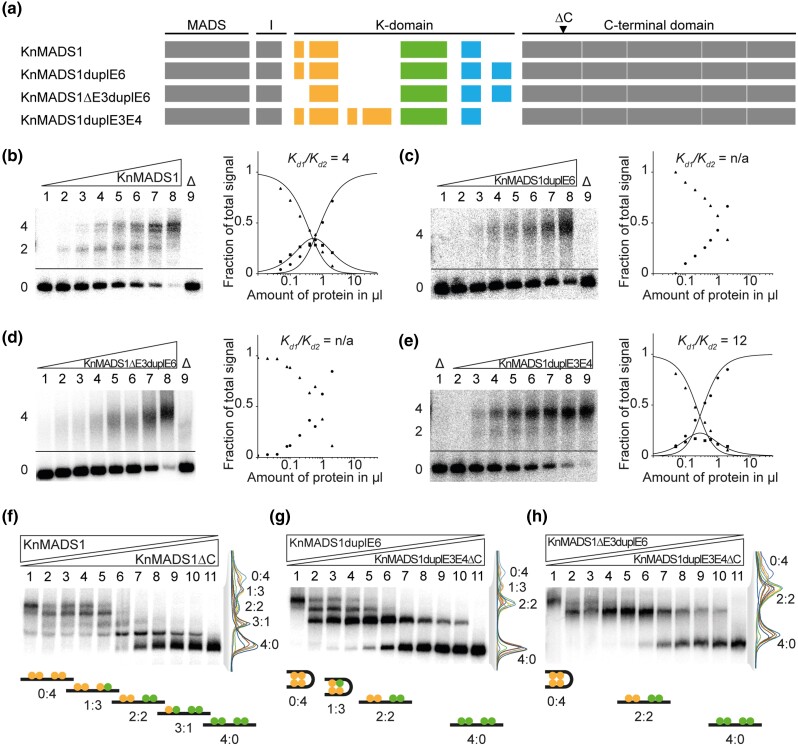
Fig. 5.
FQC formation capabilities of exon duplication and deletion mutants of the charophyte MIKC-type protein KnMADS1. (a) Exon–intron structure of KnMADS1 wild type and the mutated version KnMADS1duplE6, KnMADS1ΔE3duplE6, and KnMADS1duplE3E4. K-domain exons are color coded according to figure 3c. Black triangle highlights the position at which the coding sequence was terminated to generate C-terminally truncated versions of KnMADS1 and KnMADS1duplE3E4. (b-e) Increasing amounts of in vitro transcribed/translated (b) KnMADS1, (c) KnMADS1duplE6, (d) KnMADS1ΔE3duplE6, and (e) KnMADS1duplE3E4 was coincubated together with constant amounts of DNA probe 1. For details, see legend of figure 1. Because (c) KnMADS1duplE6 and (d) KnMADS1ΔE3duplE6 produced no signals of intermediate electrophoretic mobility constituting a DNA probe bound by a single protein dimer, Kd1/Kd2 cannot be determined. (f) KnMADS1 wild-type protein was coexpressed at different ratios with KnMADS1ΔC and coincubated together with constant amounts of DNA probe 1. An overlay of measured signal intensities of the individual lanes is shown on the right. Each peak of the graph is labeled according to the ratio of full-length and truncated protein of the corresponding fraction. The cartoons below the gel illustrate the composition of the different fractions with full-length and truncated proteins shown in yellow and green, respectively. (g, h) To test for heteromeric interaction capabilities between KnMADS1duplE6, KnMADS1ΔE3duplE6, and KnMADS1duplE3E4, the same assay as in f was conducted using (g) KnMADS1duplE6 together with KnMADS1duplE3E4ΔC and (h) KnMADS1ΔE3duplE6 together with KnMADS1duplE3E4ΔC, respectively. (b–e) Applied amounts of in vitro transcription/translation products were (lanes 1–8) 0, 0.05, 0.1, 0.2, 0.4, 0.6, 1, and 2 µl. (f–h) 3 µl of in vitro transcription/translation product were applied to each lane. Ratios of both template plasmids used for in vitro transcription/translation were (lanes 1–11): 0:1, 1:9, 1:7, 1:5, 1:3, 1:1, 3:1, 5:1, 7:1, 9:1, 1:0.