Extended Data Fig. 4.
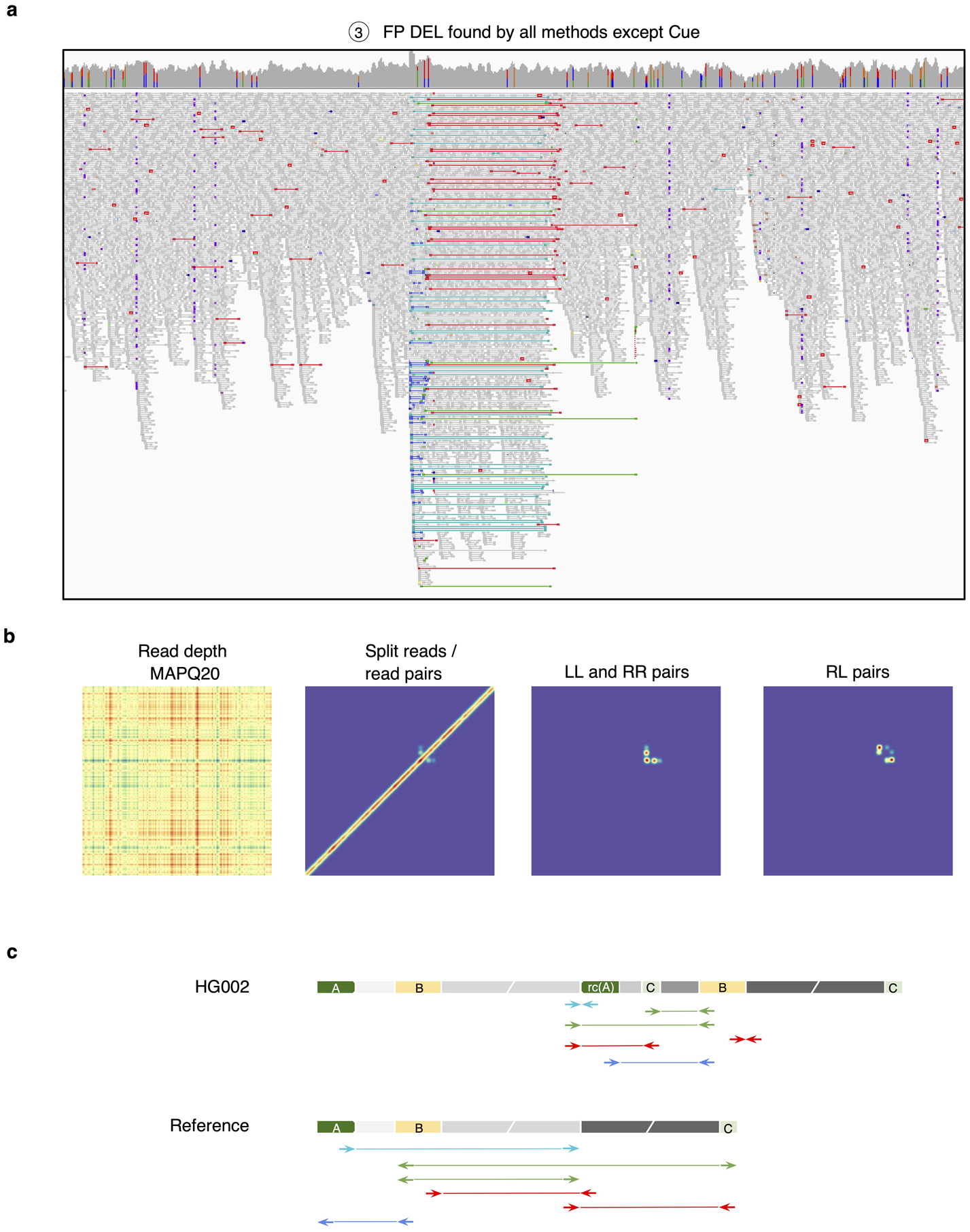
Analysis of a false positive HG002 deletion generated by all short-read callers except Cue. a. IGV plot showing short-read alignments at the call locus. Discordant read pairs mapped to the same strand (LL and RR mappings) are shown in light and dark blue, RL mappings are shown in green, and read pairs with a discordantly large insert size are shown in red. b. Cue-generated image channels depicting short-read signals that are inconsistent with a valid DEL signature. c. One of the two haplotypes of HG002, reconstructed by de novo assembly of PacBio CCS reads, that explains the main discordant pair mappings in panel a (the other haplotype is identical to the reference). The reconstructed haplotype contains two dispersed DUPs, one inverted dispersed DUP, and no DEL. Colored blocks labeled with letters are distinct short repeats. Gray blocks broken by diagonal lines are long sequences. rc(A) denotes the reverse-complement of A. Haplotypes were reconstructed and compared to the reference as follows. Let W be the sequence of the reference that covers the main patterns of discordant pairs in panel a. We built a joint de Bruijn graph (k=87) on W and on the 190 CCS reads that have some alignment to W, we removed k-mers with frequency one, and we translated W and every read into a walk (which may contain cycles) in the graph.
