Fig. 3.
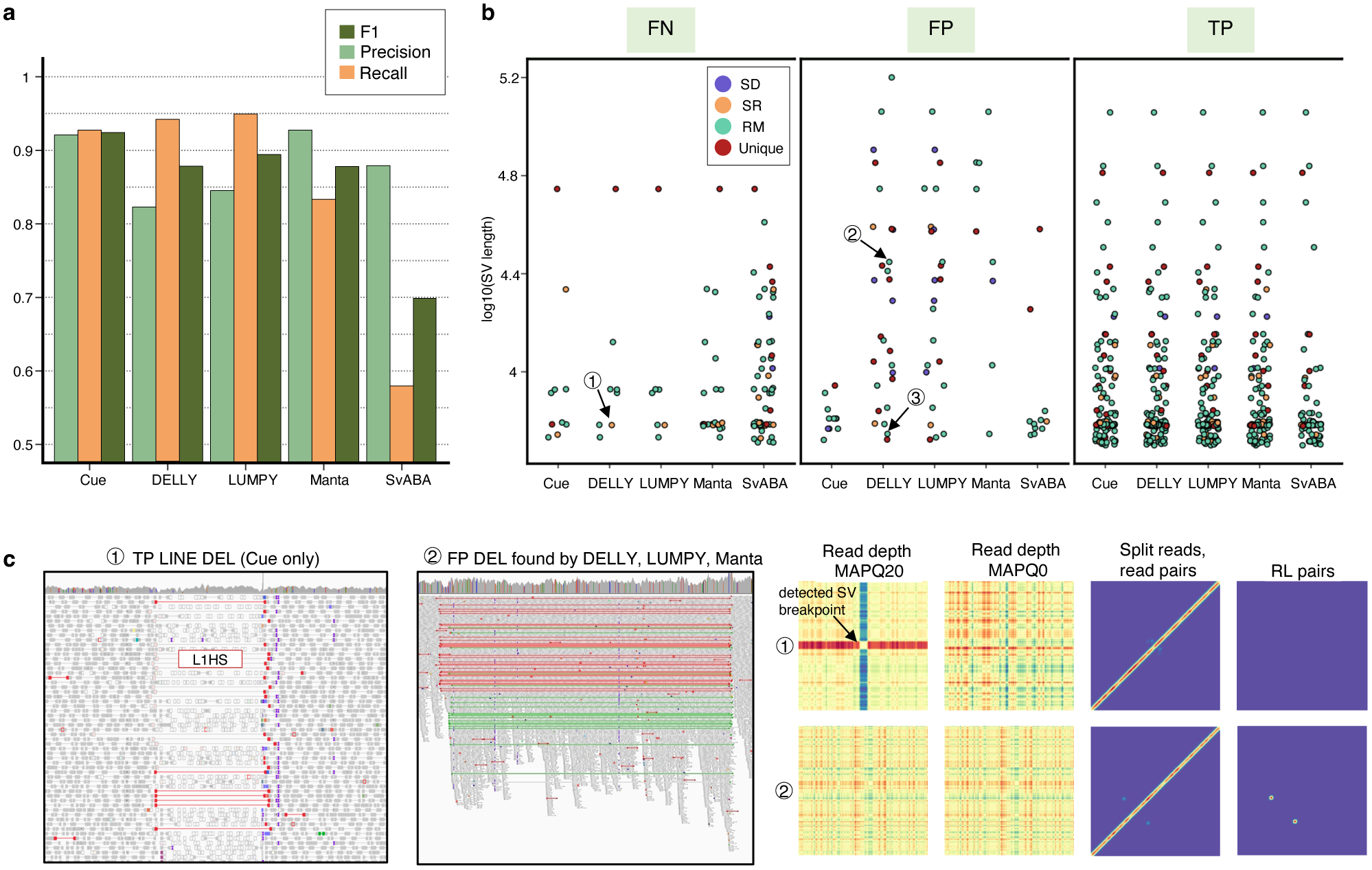
Performance evaluation on the HG002 GIAB DEL benchmark. a. Precision, recall, and F1 score in DEL calling. b. FN, FP, and TP calls broken down by size and genome context. (1)-(3) are FN and FP SV calls analyzed in panel c and Extended Data Fig. 4. c. IGV plots and Cue image channels for (1) TP LINE-1 deletion event detected only by Cue and (2) FP deletion call made by DELLY, LUMPY, and Manta. IGV shows RL read-pair alignments in green and read pairs with a discordantly large insert size in red. As we can see in the Cue-generated channels, the LINE-1 deletion signature of event (1) is well-captured in the high-MAPQ read-depth channel (which shows the drop in coverage consistent with a deletion or a repeat) and the split-read/read-pair channel (which shows the novel adjacency formed by discordant read pairs). The signatures in these two channels jointly, along with the absence of signal in the remaining channels, can uniquely characterize a deletion of a repeat element. On the other hand, while the split-read/read-pair channel alone at the site of FP event (2) can be consistent with a deletion, the presence of the RL signal and the absence of read-depth signal are not jointly consistent with a deletion. Long-read analysis found this event to be a divergent repeat as described in Extended Data Fig. 5 and Supplementary Note 2.
