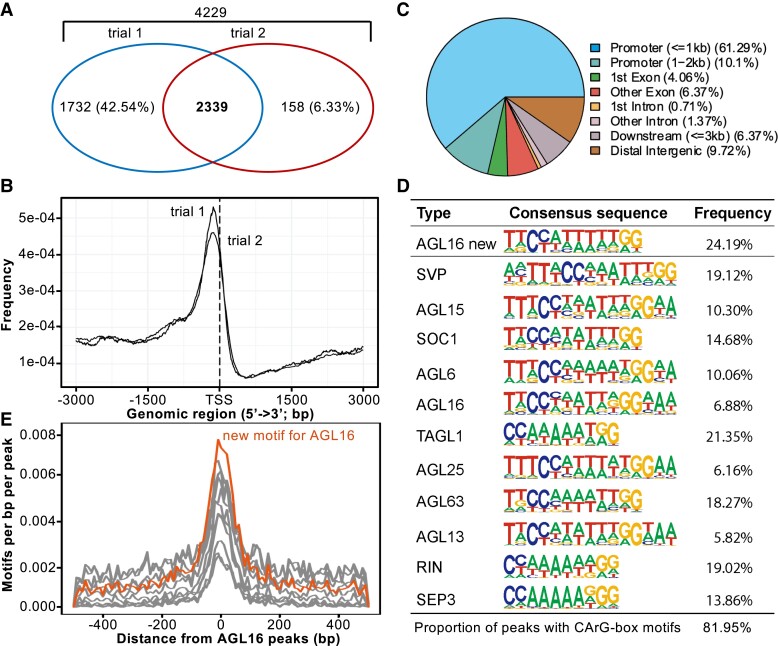
Figure 2.
Genome-wide identification of AGL16 target genes via ChIP-seq. A) Venn diagram of AGL16 targets identified in two independent trials. B) Distribution of AGL16 binding sites for two trials surrounding the transcriptional starting site (TSS). C) Location distribution in relative to nearby genes for AGL16 binding sites of trial 1. Peaks within the 3-kb promoter region were taken as AGL16 targets. D) CArG type of motifs over-represented in the AGL16 binding peaks. AGL16 new, which was highly similar to known SOC1 type, showed the de novo motif predicted for AGL16. Frequency gave the percentage for each motif presented in the binding peaks. E) Distribution of new (orange) and known (gray; shown in (D) CArG type of motifs around AGL16 peaks center.