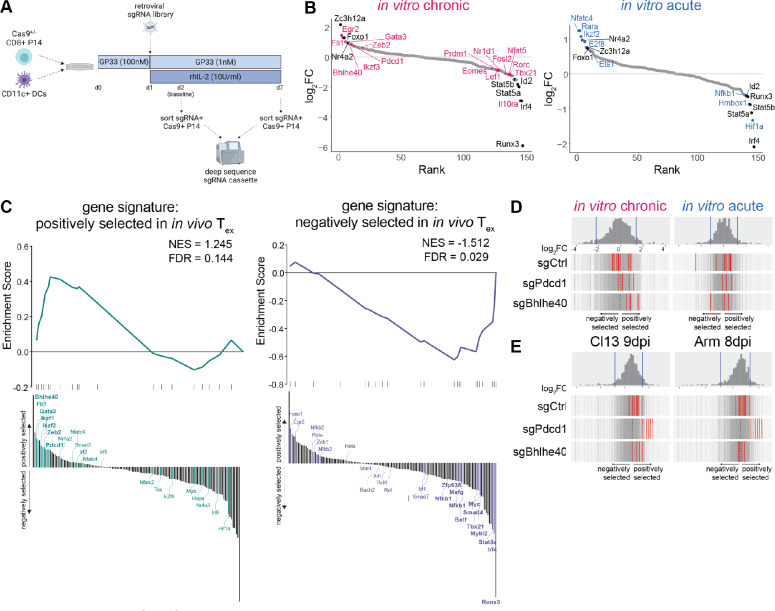
Fig. 4. Pooled CRISPR screening in in vitro chronically stimulated P14 cells identifies novel transcriptional regulators of CD8 T cell differentiation.
(A) Experiment schematic of pooled CRISPR screening in in vitro chronically stimulated P14 cells. (B) Genes targeted by pooled CRISPR screen ranked by magnitude of selection (as quantified by lfc) after acute and chronic stimulation in vitro. Colored text indicates hits unique to each condition; black text indicates hits common to both. (C) GSEA of gene sets constructed from [left] positively selected and [right] negatively selected sgRNAs from previously published pooled in vivo CRISPR screening (57). Waterfall plot illustrates rank order of hits in in vitro chronically stimulated P14 cells; bold lettering indicates leading edge genes. (D-E) Selection of individual sgRNAs against negative control genes, Pdcd1, and Bhlhe40 (D) after acute and chronic stimulation in vitro and (E) from in vivo data set (57) in PBMC at 9dpi of LCMV-Cl13 and 8dpi of LCMV-Arm. Histogram and vertical gray bars indicate distribution of all sgRNAs; vertical red bars represent indicated sgRNAs. (B-D) Representative of two experiments (two technical replicates and 2 biological replicates each).