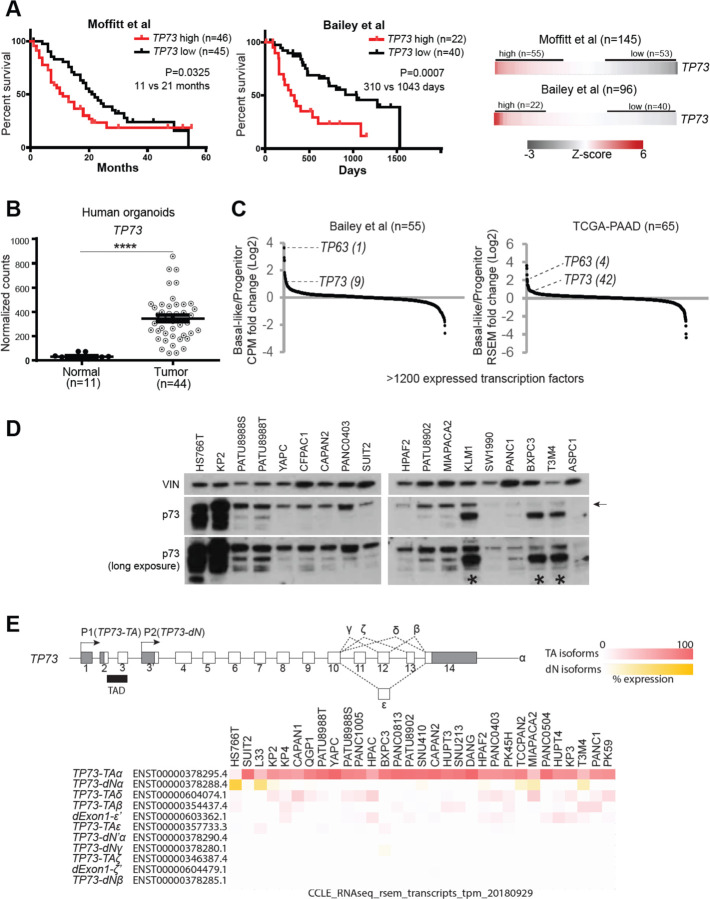
Figure 1. TP73 is prognostic, aberrantly upregulated in the PDAC tumor organoids, and preferentially expressed in the squamous subtype of PDAC.
(A) Survival curves of patients stratified by TP73 expression. Log-rank (Mantel-Cox) test used to assess the median survival values and p-values. (B) Expression of TP73 in human organoids derived from normal pancreata (Normal) or PDAC tumors (Tumor) (Tiriac et al., 2018). P-value calculated using unpaired t-test. ****, P ≤ 0.0001. Mean ± SEM shown. (C) Expression of transcription factors in the basal-like vs progenitor subtype of tumors. Transcription factors are ranked by their mean log2 fold-change in expression levels in the basal-like vs. progenitor subtype tumors. TP63, a master regulator of the squamous subtype PDAC (Somerville et al., 2018) is shown as a reference, and the rank of each gene is written inside parentheses. (D) Western blot analysis of TP73 in PDAC cell lines using a pan-TP73 antibody. VINCULIN (VIN) shown as a loading control. Bands corresponding to the molecular weight of the longest p73 isoform (i.e. p73-TAα) indicated with an arrow. Asterisk points to antibody cross-reactivity with p63-dN in cell lines expressing high levels of p63-dN. (E) (Top) Schematic of the human TP73 gene. Exons (white boxes), UTRs (gray boxes), and introns (thin lines) are depicted. Exons are numbered. Transactivation domain (TAD) is indicated with black box. 3’ spliced isoforms are indicated in Greek letters. (Bottom) TP73 isoform expression in PDAC cell lines from CCLE (Ghandi et al., 2019). Isoforms expressed from P1 or P2 promoter are referred to as TP73-TA or TP73-dN, respectively. Isoforms lacking exon1 but retaining the transactivation domain are noted as dExon1. 3’ spliced isoforms with variations at the 3’ UTR are marked with a prime (‘) symbol (i.e. dExon1-ε’ and dExon1-ζ’). Isoform expressed from an alternative promoter other than P1 or P2 is noted as dN’ (i.e. TP73- dN’α). For the heatmap, cell lines are listed in descending order (from left to right) of total TP73 isoform expression. TP73-TA and dExon1 are collectively considered as TA isoforms, and TP73-dN and TP73- dN’ are collectively considered as dN isoforms. Expression of each isoform is normalized within a cell line (i.e. % expression = tpm value of isoform A / sum of tpm values of all isoforms * 100).