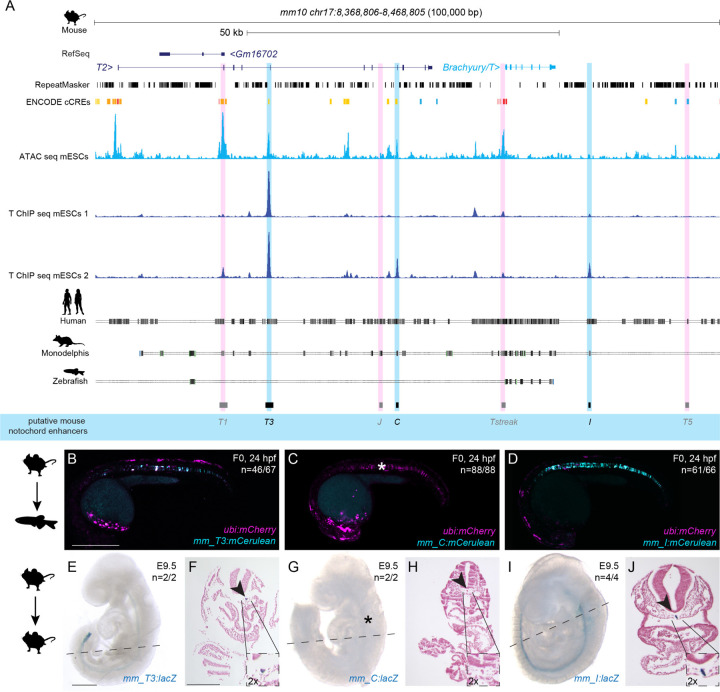
Figure 3: Mouse Brachyury enhancer elements are active in different species.
(A) Mouse Brachyury/T/TBXTB locus adapted from the UCSC genome browser. Repeats marked in black using the RepeatMasker track. Further annotated is the track with ENCODE conserved cis-regulatory elements (cCREs). ATAC-sequencing (light blue peaks) and T ChIP-sequencing (dark blue lines) indicate enhancer elements (light pink highlight, not active; light blue highlight, active) that are conserved in human and Monodelphis.
(B,C,D) Representative F0 zebrafish embryos injected with the mouse enhancer elements mm_T3 (B), mm_C (C), and mm_I (D). mm_T3 and mm_I show mosaic mCerulean reporter expression in the notochord at 24 hpf and mosaic ubi:mCherry expression as injection control. Mouse enhancer element mm_C is not active in the zebrafish notochord (asterisk in C). N represent the number of animals expressing mCerulean in the notochord relative to the total number of animals expressing mCherry. Scale bar in B: 0.5 mm, applies to B-D.
(E,G,I) Representative images of transgenic E9.5 mouse embryos expressing lacZ (encoding beta-galactosidase) under the mouse enhancer elements mm_T3 (E), mm_C (G) and mm_I (I) visualized with X-gal whole mount staining. While mm_T3 and mm_I express beta-galactosidase in the entire notochord, beta-galactosidase expression from mouse mm_C is absent (asterisk in G). N represent the number of animals expressing beta-galactosidase in the notochord relative to the total number of animals with tandem integrations at H11. Dotted lines represent the sectioning plane. Scale bar in E: 0.5 mm, applies to E,G,I.
(F,H,J) Representative images of Fast Red-stained cross sections from embryos shown on the left, mm_T3 (F), mm_C (H), and mm_I (J). Black arrowheads point at notochord, and inserts show notochords at 2x higher magnification. Scale bar in F: 0.25 mm, applies to F,H,J.