Abstract
While large language models (LLMs) have been successfully applied to various tasks, they still face challenges with hallucinations and generating erroneous content. Augmenting LLMs with domain-specific tools such as database utilities has the potential to facilitate more precise and straightforward access to specialized knowledge. In this paper, we present GeneGPT, a novel method for teaching LLMs to use the Web Application Programming Interfaces (APIs) of the National Center for Biotechnology Information (NCBI) and answer genomics questions. Specifically, we prompt Codex (code–davinci–002) to solve the Gene Turing tests with few-shot URL requests of NCBI API calls as demonstrations for in-context learning. During inference, we stop the decoding once a call request is detected and make the API call with the generated URL. We then append the raw execution results returned by NCBI APIs to the generated texts and continue the generation until the answer is found or another API call is detected. Our preliminary results show that GeneGPT achieves state-of-the-art results on three out of four one-shot tasks and four out of five zero-shot tasks in the Gene Turing dataset. Overall, GeneGPT achieves a macro-average score of 0.76, which is much higher than retrieval-augmented LLMs such as the New Bing (0.44), biomedical LLMs such as BioMedLM (0.08) and BioGPT (0.04), as well as other LLMs such as GPT-3 (0.16) and ChatGPT (0.12).
1. Introduction
Large language models (LLMs) such as PaLM (Chowdhery et al., 2022) and GPT-4 (OpenAI, 2023) have shown great success on a wide range of general-domain Natural Language Processing (NLP) tasks. They also achieve state-of-the-art performance on many domain-specific tasks like biomedical question answering (Singhal et al., 2022; Liévin et al., 2022; Nori et al., 2023). However, since there is no intrinsic mechanism for auto-regressive LLMs to “consult” with any source of truth, they can generate plausible-sounding but incorrect content (Ji et al., 2023). To tackle the hallucination issue, various studies have been proposed to augment LLMs (Mialon et al., 2023) by either conditioning them on retrieved relevant content (Guu et al., 2020; Lewis et al., 2020; Borgeaud et al., 2022) or allowing them to use other external tools such as program APIs (Parisi et al., 2022; Schick et al., 2023; Qin et al., 2023).
In this work, we propose to teach LLMs to use the Web APIs1 of the National Center for Biotechnology Information (NCBI). NCBI provides API access to its biomedical databases and tools including Entrez Programming Utilities (E-utils) and Basic Local Alignment Search Tool (BLAST) URL API (Altschul et al., 1990; Schuler et al., 1996; Sayers et al., 2019). Enabling LLMs to use NCBI Web APIs can provide easier and more precise access to biomedical information, especially for users who are inexperienced with the database systems. The advantage of Web API is to relieve users from implementing functionalities, maintaining large databases, and heavy computation burdens because the only requirement is an internet connection.
We introduce GeneGPT, a novel method that prompts Codex (Chen et al., 2021) to use NCBI Web APIs by in-context learning (Brown et al., 2020). GeneGPT consists of two main modules: (a) a specifically designed prompt that consists of API usage demonstrations, and (b) an inference algorithm that integrates API calls in the Codex decoding process. We evaluate GeneGPT on the Gene Turing dataset (Hou and Ji, 2023), a question answering (QA) benchmark for genomics, and compare it to a variety of other LLMs such as the New Bing2, ChatGPT3, and BioGPT (Luo et al., 2022). GeneGPT achieves the best performance on three out of four one-shot tasks and four out of five zero-shot tasks, where one instance and no instance are included in the prompt, respectively. It also achieves the second-highest result on the rest tasks. On average, GeneGPT scores 0.76, which is much higher than the previous SOTA (0.44 by New Bing). We hope this pilot study can provide insights for developing systems that improve biomedical information access by integrating LLMs with domain-specific tools such as NCBI Web APIs.
2. GeneGPT
In this section, we first introduce the general syntax of NCBI Web APIs (§2.1). We then describe two key components of GeneGPT, including its prompt design (§2.2) and the inference algorithm (§2.3) for downstream tasks.
2.1. NCBI Web APIs
We utilize NCBI Web APIs of E-utils that provide access to genomics databases and the BLAST tool for DNA sequence alignment. All Web API calls are implemented by the urllib library in Python.
E-utils is the API for accessing the Entrez system (Schuler et al., 1996), a database system that covers 38 NCBI databases of biomedical data (Sayers et al., 2019), such as nucleotide and protein sequences. It provides a fixed URL syntax for rapidly retrieving biomedical information. Specifically, the base URL for an E-utils request is “https://eutils.ncbi.nlm.nih.gov/entrez/eutils/{function}.fcgi”, where function can be esearch, efetch, esummary and etc. esearch returns the unique database identifiers for a given query term, while efetch and esummary return specific information for a given list of identifiers. Important arguments in the URL request include the search term or ids (term or id), the database to use (db), the maximum number of returned items (retmax), and the return format (retmode).
BLAST URL API allows users to submit queries to find regions of similarities between nucleotide or protein sequences to existing databases using the BLAST algorithm (Altschul et al., 1990; Boratyn et al., 2013) on NCBI servers. The results can be used to infer relationships between sequences or identify members of gene families. The base URL for the BLAST URL API is “https://blast.ncbi.nlm.nih.gov/blast/Blast.cgi”. By sending different parameters to this API, user can submit and retrieve queries that are executed by NCBI web servers. Every call to the API must include a CMD parameter that defines the type of the call. When submitting queries using CMD=Put, the user can specify the querying database with the DATABASE parameter, the searching program with the PROGRAM parameter, and the query sequence with the QUERY parameter. The user will get an RID after the CMD=Put API call, and can make another API call with the Get command and the returned RID to retrieve its BLAST results. More details can be found in the NCBI BLAST API documentation4.
2.2. Prompt design
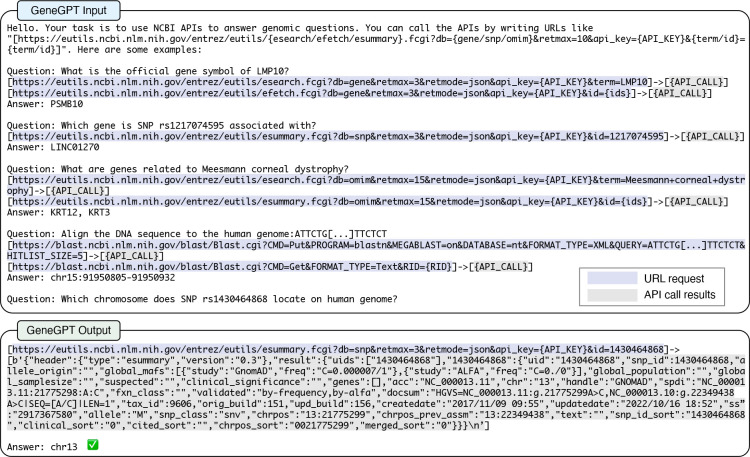
Figure 1 shows an example of our prompt, which consists of three parts. The first two parts are universal across different tasks, and the last part includes a specific test question for inference:
Figure 1:
An example of teaching large language models to use NCBI Web APIs and solve genomics questions. The prompt includes three parts: 1. a general task description and an NCBI Web API URL template; 2. Four demonstrations of NCBI API usage (summarized in Table 1); 3. A task-specific test question.
-
1
The prompt starts with an overall task description (“Your task is to use NCBI APIs to answer genomics questions.”) and the NCBI Web API URL template (described in §2.1).
-
2
It is followed by four QA instances as demonstrations of using NCBI Web APIs, which are summarized in Table 1. We use them to teach the LLM to use three functions (esearch, efetch, esummary) and three databases (gene, snp, omim) of the NCBI E-utils, as well as the BLAST API. The API URLs and the call results are marked up by “[ ]”, with a special “–>” symbol inserted in between.
Table 1:
Summary of in-context demonstrations of NCBI Web API usage. We use the QA instances from four tasks: gene alias, gene SNP association, gene disease association, and DNA sequence alignment to human genome (described in §3.1).
| # | Task | Database | Function |
|---|---|---|---|
|
| |||
| 1 | Alias | gene | esearch, efetch |
| 2 | Gene SNP | snp | esummary |
| 3 | Gene disease | omim | esearch, esummary |
| 4 | Alignment | nt | blastn |
Algorithm 1.
GeneGPT inference algorithm
| Input: question |
| Model: Codex (code-davinci-002) |
| Output: answer |
| prompt ← header + demonstrations + question |
| finished ← False |
| while not finished do |
| next token ← Codex(prompt) |
| prompt ← prompt + next token |
| if next token is “–>” then |
| # help call Web API |
| url ← extractLastURL(prompt) |
| result ← callWebAPI(url) |
| # append API call result |
| prompt ← prompt + result |
| else if next token is “\n\n” then |
| answer ← extractAnswer(prompt) |
| finished ← True |
| end if |
| end while |
-
3
The specific test question is then appended to the end of the prompt, with a similar format to the demonstration instances for in-context learning (Brown et al., 2020).
2.3. Inference algorithm
The GeneGPT inference algorithm is briefly shown in Algorithm 1. Specifically, we first append the given question to the prompt (described in §2.2) and feed the concatenated text to Codex (code–davinci–002, Chen et al. (2021)) with a temperature of 0. We choose to use Codex for two reasons: (1) it is pre-trained with code data and shows better code understanding abilities, which is crucial in generating the URLs and interpreting the raw API results; (2) its API has the longest (8k tokens) input length among all available models so that we can fit the demonstrations in.
We discontinue the text generation process when the special “–>” symbol is detected, which is the indication for an API call request. Then we extract the last URL and call the NCBI Web API with it. The raw execution results will be appended to the generated text, and it will be fed to Codex to continue the generation. When “\n\n”, an answer indicator used in the demonstrations, is generated, we will stop the inference and extract the answer after the generated “Answer: ”.
3. Experiments
3.1. The GeneTuring dataset
The Gene Turing dataset (Hou and Ji, 2023) contains 12 tasks, and each task has 50 question-answer pairs. The tasks are classified into four categories: nomenclature, genomics location, functional analysis, and sequence alignment. We use 9 GeneTuring tasks that are related to NCBI resources to evaluate the proposed GeneGPT method. The chosen tasks are briefly described below5:
Nomenclature is about gene names. We use the gene alias task and the gene name conversion task, where the objective is to find the official gene symbols for their non-official synonyms.
genomics location is about the locations of genes, single-nucleotide polymorphism (SNP), and their relations. We include the gene location, SNP location, and gene SNP association tasks. The first two tasks ask for the chromosome locations (e.g., “chr2”) of a gene or an SNP, and the last one asks for related genes for a given SNP.
Functional analysis is about gene functions. We use the gene disease association task where the goal is to return related genes for a given disease, and the protein-coding genes task which asks whether a gene is a protein-coding gene or not.
Sequence alignment is about DNA sequences. We use the DNA sequence alignment to human genome task and the DNA sequence alignment to multiple species task. The former maps an DNA sequence to a specific human chromosome, while the latter maps an DNA sequence to a specific species (e.g. “zebrafish”).
3.2. Compared methods
We compare the proposed GeneGPT method with various baselines evaluated by Hou and Ji (2023), including general-domain GPT-based (Radford et al., 2018) LLMs such as GPT-2 (Radford et al., 2019), GPT-36 (Brown et al., 2020), and ChatGPT7, GPT-2-sized biomedical domain-specific LLMs such as BioGPT (Luo et al., 2022) and BioMedLM8 (previously known as PubMedGPT), as well as the New Bing9, a retrieval-augmented LLM that has access to relevant web pages retrieved by the Bing search engine.
3.3. Evaluation
For the performance of the compared methods, we directly use the results reported in Hou and Ji (2023) that are manually evaluated.
To evaluate our proposed GeneGPT method, we follow the general criteria but perform automatic evaluations. Specifically, we only consider exact matches between model predictions and the ground truth as correct predictions for all nomenclature and genomics location tasks. For the gene disease association task, we measure the recall as in Hou and Ji (2023) but based on exact individual gene matches. For the protein-coding genes task and the DNA sequence alignment to multiple species task, we also consider exact matches as correct after applying a simple vocabulary mapping that converts model-predicted “yes”/“no” to “TRUE”/“NA” and Latin species names to their informal names (e.g., “Saccharomyces cerevisiae” to “yeast”), respectively. For the DNA sequence alignment to human genome task, we give correct chromosome mapping but incorrect position mapping a score of 0.5 (e.g., chr8:7081648–7081782 v.s. chr8:1207812–1207946), since the original task does not specify a reference genome. Overall, our evaluation of GeneGPT is more strict than the original evaluation of other LLMs in Hou and Ji (2023), which performs manual evaluation and might consider non-exact matches as correct.
3.4. Main results
Table 2 shows the performance of GeneGPT on the GeneTuring tasks in comparison with other LLMs. For GeneGPT, four tasks (with “*” in Table 2) are one-shot where one instance is used for API demonstration, and the other five tasks are zero-shot. For the compared LLMs, all tasks are zero-shot.
Table 2:
Performance of GeneGPT compared to other LLMs on the GeneTuring dataset.
| GeneTuring task | GET-2 | BioGPT | BioMedLM | GET-3 | ChatGPT | New Bing | GeneGPT |
|---|---|---|---|---|---|---|---|
|
| |||||||
| Nomenclature | |||||||
| Gene alias* | 0.00 | 0.00 | 0.04 | 0.09 | 0.07 | 0.66 | 0.80 |
| Gene name conversion | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.85 | 0.98 |
| Average | 0.00 | 0.00 | 0.02 | 0.05 | 0.04 | 0.76 | 0.89 |
|
| |||||||
| genomics location | |||||||
| Gene SNP association* | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 1.00 |
| Gene location | 0.01 | 0.04 | 0.12 | 0.09 | 0.09 | 0.61 | 0.62 |
| SNP location | 0.03 | 0.05 | 0.01 | 0.02 | 0.05 | 0.01 | 1.00 |
| Average | 0.01 | 0.03 | 0.04 | 0.04 | 0.05 | 0.21 | 0.87 |
|
| |||||||
| Functional analysis | |||||||
| Gene disease association* | 0.00 | 0.02 | 0.16 | 0.34 | 0.31 | 0.84 | 0.49 |
| Protein-coding genes | 0.00 | 0.18 | 0.37 | 0.70 | 0.54 | 0.97 | 0.66 |
| Average | 0.00 | 0.10 | 0.27 | 0.52 | 0.43 | 0.91 | 0.58 |
|
| |||||||
| Sequence alignment | |||||||
| DNA to human genome* | 0.02 | 0.07 | 0.03 | 0.00 | 0.00 | 0.00 | 0.44 |
| DNA to multiple species | 0.02 | 0.00 | 0.00 | 0.20 | 0.00 | 0.00 | 0.86 |
| Average | 0.02 | 0.04 | 0.02 | 0.10 | 0.00 | 0.00 | 0.65 |
|
| |||||||
| Overall average | 0.00 | 0.04 | 0.08 | 0.16 | 0.12 | 0.44 | 0.76 |
One-shot learning for GeneGPT, where one instance is included in the prompt for NCBI Web API demonstration. Unlabeled tasks are zero-shot for GeneGPT. Bolded numbers denote the highest performance, while underlined numbers denote the second-highest performance.
Nomenclature:
GeneGPT achieves state-of-the-art (SOTA) performance on both the one-shot gene alias task (an accuracy of 0.80) and the zero-shot gene name conversion task (an accuracy of 0.98). On average, GeneGPT outperforms New Bing by a large margin (0.89 v.s. 0.76). All other GPT models have accuracy scores of less than 0.10 on the nomenclature tasks.
genomics location:
GeneGPT also achieves SOTA performance on all genomics location tasks, including the one-shot gene SNP association task (1.00 accuracy), as well as the zero-shot gene location task (0.62 accuracy) and the zero-shot SNP location task (1.00 accuracy). While the New Bing is comparable to GeneGPT on gene location (0.61 v.s. 0.62), its performance on the two SNP-related tasks is close to 0. Similarly, most other LLMs score less than 0.10.
Functional analysis:
New Bing performs better functional analysis tasks than the proposed GeneGPT (average score: 0.91 v.s. 0.58), which is probably because many web pages related to gene functions can be retrieved by the Bing search engine. We also note that other LLMs, especially GPT-3 and ChatGPT, perform moderately well and much better than they perform on other tasks. This might also be due to the fact that many gene-function-related texts are included in their pre-training corpora.
Sequence alignment:
GeneGPT performs much better with an average score of 0.65 than all other models including New Bing (0.00), which essentially fails on the sequence alignment tasks. This is not very surprising since sequence alignment is easy with the BLAST tool, but almost impossible for an auto-regressive LLM even with retrieval augmentation as the input sequences are too specific to appear on any web pages.
Although evaluated under a more strict setting (§3.3), GeneGPT achieves a macro-average performance of 0.76 which is much higher than other compared LLMs including New Bing (0.44). Overall, GeneGPT achieves new SOTA performance on three out of four one-shot tasks and four out of five zero-shot tasks and is outperformed by New Bing only on the 2 functional analysis tasks.
4. Conclusions
We present GeneGPT, a novel method that teaches large language models to use NCBI Web APIs by in-context learning. Preliminary results show that GeneGPT achieves state-of-the-art performance on 7 GeneTuring tasks, in comparison to various LLMs including New Bing. This indicates that external tools might be superior to relevant web pages for augmenting LLMs to solve genomics questions.
We plan to extend this pilot study with two future directions: (1) fine-tuning LLMs with NCBI API calls instead of in-context learning and (2) exploring multi-hop biomedical question answering (Jin et al., 2022) and chain-of-thought prompting (Wei et al., 2022) to better serve real-life information needs about biomedicine.
Acknowledgements
We are grateful to the GeneTuring authors for sharing the dataset. We also thank the NCBI API support team for helpful discussions. This research was supported by the NIH Intramural Research Program, National Library of Medicine. Qingyu Chen was also supported by the National Library of Medicine of the National Institutes of Health under award number 1K99LM014024.
Footnotes
We refer our readers to Hou and Ji (2023) for more details.
Specifically GPT-3.5 (text-davinci-003).
https://chat.openai.com/ (Jan 31 version).
References
- Altschul Stephen F, Gish Warren, Miller Webb, Myers Eugene W, and Lipman David J. 1990. Basic local alignment search tool. Journal of molecular biology, 215(3):403–410. [DOI] [PubMed] [Google Scholar]
- Boratyn Grzegorz M, Camacho Christiam, Cooper Peter S, Coulouris George, Fong Amelia, Ma Ning, Madden Thomas L, Matten Wayne T, McGinnis Scott D, Merezhuk Yuri, et al. 2013. Blast: a more efficient report with usability improvements. Nucleic acids research, 41(W1):W29–W33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgeaud Sebastian, Mensch Arthur, Hoffmann Jordan, Cai Trevor, Rutherford Eliza, Millican Katie, Van Den Driessche George Bm, Lespiau Jean-Baptiste, Damoc Bogdan, Clark Aidan, et al. 2022. Improving language models by retrieving from trillions of token s. In International conference on machine learning, pages 2206–2240. PMLR. [Google Scholar]
- Brown Tom, Mann Benjamin, Ryder Nick, Subbiah Melanie, Jared D Kaplan Prafulla Dhariwal, Neelakantan Arvind, Shyam Pranav, Sastry Girish, Askell Amanda, et al. 2020. Language models are few-shot learners. Advances in neural information processing systems, 33:1877–1901. [Google Scholar]
- Chen Mark, Tworek Jerry, Jun Heewoo, Yuan Qiming, de Oliveira Pinto Henrique Ponde, Kaplan Jared, Edwards Harri, Burda Yuri, Joseph Nicholas, Brockman Greg, et al. 2021. Evaluating large language models trained on code. arXiv preprint arXiv:2107.03374. [Google Scholar]
- Chowdhery Aakanksha, Narang Sharan, Devlin Jacob, Bosma Maarten, Mishra Gaurav, Roberts Adam, Barham Paul, Won Chung Hyung, Sutton Charles, Gehrmann Sebastian, et al. 2022. Palm: Scaling language modeling with pathways. arXiv preprint arXiv:2204.02311. [Google Scholar]
- Guu Kelvin, Lee Kenton, Tung Zora, Pasupat Panupong, and Chang Mingwei. 2020. Retrieval augmented language model pre-training. In International conference on machine learning, pages 3929–3938. PMLR. [Google Scholar]
- Hou Wenpin and Ji Zhicheng. 2023. Geneturing tests gpt models in genomics. bioRxiv, pages 2023–03. [Google Scholar]
- Ji Ziwei, Lee Nayeon, Frieske Rita, Yu Tiezheng, Su Dan, Xu Yan, Ishii Etsuko, Bang Ye Jin, Madotto Andrea, and Fung Pascale. 2023. Survey of hallucination in natural language generation. ACM Computing Surveys, 55(12):1–38. [Google Scholar]
- Jin Qiao, Yuan Zheng, Xiong Guangzhi, Yu Qianlan, Ying Huaiyuan, Tan Chuanqi, Chen Mosha, Huang Songfang, Liu Xiaozhong, and Yu Sheng. 2022. Biomedical question answering: a survey of approaches and challenges. ACM Computing Surveys (CSUR), 55(2):1–36. [Google Scholar]
- Lewis Patrick, Perez Ethan, Piktus Aleksandra, Petroni Fabio, Karpukhin Vladimir, Goyal Naman, Heinrich Küttler Mike Lewis, Yih Wen-tau, Rocktäschel Tim, et al. 2020. Retrieval-augmented generation for knowledge-intensive nlp tasks. Advances in Neural Information Processing Systems, 33:9459–9474. [Google Scholar]
- Valentin Liévin Christoffer Egeberg Hother, and Winther Ole. 2022. Can large language models reason about medical questions? arXiv preprint arXiv:2207.08143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Renqian, Sun Liai, Xia Yingce, Qin Tao, Zhang Sheng, Poon Hoifung, and Liu Tie-Yan. 2022. Biogpt: generative pre-trained transformer for biomedical text generation and mining. Briefings in Bioinformatics, 23(6). [DOI] [PubMed] [Google Scholar]
- Mialon Grégoire, Dessì Roberto, Lomeli Maria, Nalmpantis Christoforos, Pasunuru Ram, Raileanu Roberta, Baptiste Rozière Timo Schick, Dwivedi-Yu Jane, Celikyilmaz Asli, et al. 2023. Augmented language models: a survey. arXiv preprint arXiv:2302.07842. [Google Scholar]
- Nori Harsha, King Nicholas, McKinney Scott Mayer, Carignan Dean, and Horvitz Eric. 2023. Capabilities of gpt-4 on medical challenge problems. arXiv preprint arXiv:2303.13375. [Google Scholar]
- OpenAI. 2023. GPT-4 technical report. CoRR, abs/2303.08774. [Google Scholar]
- Parisi Aaron, Zhao Yao, and Fiedel Noah. 2022. Talm: Tool augmented language models. arXiv preprint arXiv:2205.12255. [Google Scholar]
- Qin Yujia, Hu Shengding, Lin Yankai, Chen Weize, Ding Ning, Cui Ganqu, Zeng Zheni, Huang Yufei, Xiao Chaojun, Han Chi, et al. 2023. Tool learning with foundation models. arXiv preprint arXiv:2304.08354. [Google Scholar]
- Radford Alec, Narasimhan Karthik, Salimans Tim, Sutskever Ilya, et al. 2018. Improving language understanding by generative pre-training. [Google Scholar]
- Radford Alec, Wu Jeffrey, Child Rewon, Luan David, Amodei Dario, Sutskever Ilya, et al. 2019. Language models are unsupervised multitask learners. OpenAI blog, 1(8):9. [Google Scholar]
- Sayers Eric W, Agarwala Richa, Bolton Evan E, Brister J Rodney, Canese Kathi, Clark Karen, Connor Ryan, Fiorini Nicolas, Funk Kathryn, Hefferon Timothy, et al. 2019. Database resources of the national center for biotechnology information. Nucleic acids research, 47(Database issue):D23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schick Timo, Dwivedi-Yu Jane, Dessì Roberto, Raileanu Roberta, Lomeli Maria, Zettlemoyer Luke, Cancedda Nicola, and Scialom Thomas. 2023. Tool-former: Language models can teach themselves to use tools. arXiv preprint arXiv:2302.04761. [Google Scholar]
- Schuler GD, Epstein JA, Ohkawa H, and Kans JA. 1996. Entrez: molecular biology database and retrieval system.Methods in enzymology, 266:141–162. [DOI] [PubMed] [Google Scholar]
- Singhal Karan, Azizi Shekoofeh, Tu Tao, Mahdavi S Sara, Wei Jason, Chung Hyung Won, Scales Nathan, Tanwani Ajay, Cole-Lewis Heather, Pfohl Stephen, et al. 2022. Large language models encode clinical knowledge. arXiv preprint arXiv:2212.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Jason, Wang Xuezhi, Schuurmans Dale, Bosma Maarten, Chi Ed, Le Quoc, and Zhou Denny. 2022. Chain of thought prompting elicits reasoning in large language models. arXiv preprint arXiv:2201.11903. [Google Scholar]