Abstract
Aims
The present study aims to characterize the genetic risk architecture of bicuspid aortic valve (BAV) disease, the most common congenital heart defect.
Methods and results
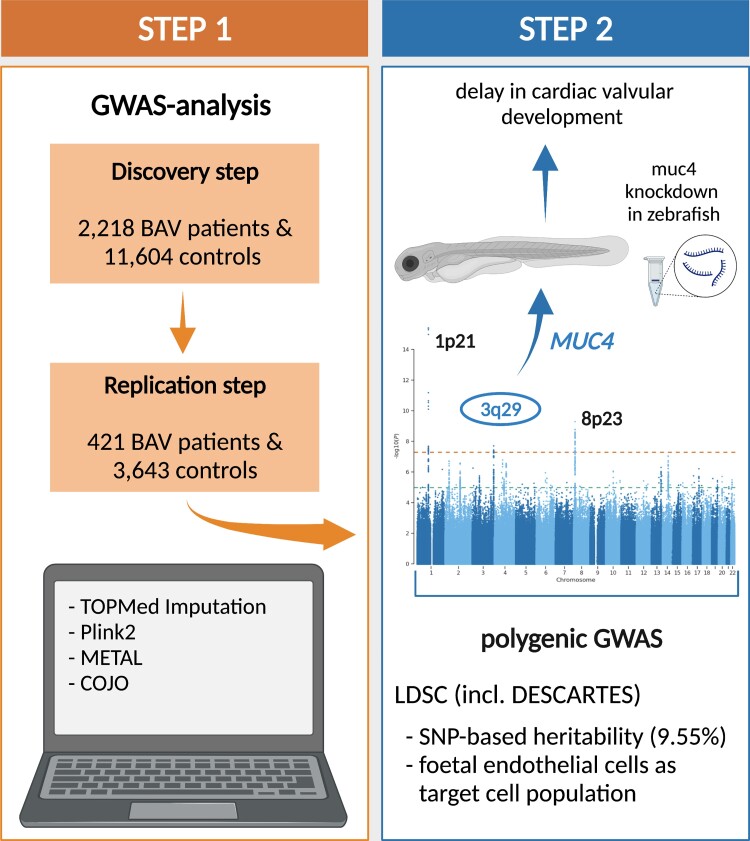
We carried out a genome-wide association study (GWAS) including 2236 BAV patients and 11 604 controls. This led to the identification of a new risk locus for BAV on chromosome 3q29. The single nucleotide polymorphism rs2550262 was genome-wide significant BAV associated (P = 3.49 × 10−08) and was replicated in an independent case–control sample. The risk locus encodes a deleterious missense variant in MUC4 (p.Ala4821Ser), a gene that is involved in epithelial-to-mesenchymal transformation. Mechanistical studies in zebrafish revealed that loss of Muc4 led to a delay in cardiac valvular development suggesting that loss of MUC4 may also play a role in aortic valve malformation. The GWAS also confirmed previously reported BAV risk loci at PALMD (P = 3.97 × 10−16), GATA4 (P = 1.61 × 10−09), and TEX41 (P = 7.68 × 10−04). In addition, the genetic BAV architecture was examined beyond the single-marker level revealing that a substantial fraction of BAV heritability is polygenic and ∼20% of the observed heritability can be explained by our GWAS data. Furthermore, we used the largest human single-cell atlas for foetal gene expression and show that the transcriptome profile in endothelial cells is a major source contributing to BAV pathology.
Conclusion
Our study provides a deeper understanding of the genetic risk architecture of BAV formation on the single marker and polygenic level.
Keywords: Bicuspid aortic valve, GWAS, SNP-based heritability, Foetal heart transcriptome, Zebrafish
Graphical Abstract
Graphical Abstract.
Time of primary review: 34 days
1. Introduction
Bicuspid aortic valve (BAV) is the most common congenital heart defect (CHD) with a prevalence of 0.5–2%.1 BAV is of major clinical relevance because as many as 71% of patients develop aortic valve stenosis (AS) and ∼60% experience aortic root dilatation with risk of aortic dissection. Furthermore, 10–30% of patients with BAV develop infective endocarditis.2 As a consequence, BAV accounts for nearly 50% of all aortic valve replacements.3
Genetic factors are strongly involved in the aetiology of BAV with an observed heritability of 47–89%.4,5 This range of heritability reflects the genetically heterogeneous nature of BAV with a wide spectrum of disease-causing mutations and risk variants. On one end of this spectrum, BAV is the result of highly penetrant mutations in single genes. This refers to monogenic BAV forms, which can be syndromic or non-syndromic. Examples of syndromic BAV that are characterized by additional extra-cardiac conditions include Loeys-Dietz syndrome with mutations in TGFBR1 or TGFBR2 as well as familial thoracic aortic aneurysm and dissection (TAAD) syndrome with mutations in ACTA2.6,7 In contrast, mutations in NOTCH1, ADAMTS19, SMAD6, or ROBO4 were found in monogenic non-syndromic cases of BAV.8–11 However, the vast majority of BAV cases are sporadic12 and result from a complex genetic background.
Genome-wide association studies (GWAS) provides an unbiased approach for discovering genetic risk variants for complex genetic traits and two GWAS involving BAV patients have been published so far. The first GWAS analysed 466 European BAV cases as discovery sample and 1326 European BAV cases as replication cohort.13 This study identified one risk single nucleotide polymorphism (SNP) at the 3′ end of GATA4 with genome-wide significant BAV association as well as one independent SNP within GATA4 that represents a missense variant (p.Ser377Gly) and was suggestive BAV associated. In addition, a rare missense variant (p.Thr1221Met) within DHX38 showed genome-wide significant BAV association. In a second GWAS of AS, the authors used a follow-up sample of 1555 European BAV patients in order to assess whether associated AS risk variants occurred on the background of a tricuspid or bicuspid valve formation.14 Of all genome-wide significant associated AS variants, SNPs at PALMD, TEX41, or MYH6 were also associated with BAV. All association findings along with information on the identified BAV risk variants and used GWAS samples are summarized in Supplementary material online, Table S1.
In the present study, we further characterized the genetic architecture of BAV and present single marker and polygenic association findings from the largest GWAS analysed to date. In addition, we functionally characterized a new BAV GWAS locus in zebrafish using CRISPR/Cas9-mediated knockout and antisense oligonucleotide Morpholino- (MO-) mediated gene knockdown.
2. Methods
2.1. Study subjects
The GWAS sample consisted of two European case–control cohorts that were recruited in Germany and the UK. The German cohort comprised 1539 BAV patients that were recruited by the University Hospitals of Bonn and Lübeck as well as the German Heart Centre Munich and 4414 controls that were part of the population-based Heinz-Nixdorf Recall study.15 The UK cohort comprised 697 BAV patients that were recruited by the University Hospitals of Leicester as well as 7190 controls that were part of the UK Biobank.16 In total, we analysed 2236 BAV patients (583 females, 1653 males) and 11 604 controls (4415 females, 7189 males) for the GWAS. The replication sample consisted of 421 German BAV patients (99 females, 322 males) that were recruited by the University Hospital of Bonn and the German Heart Centre Munich as well as 3643 controls (1266 females, 2377 males) that were part of a blood donor cohort recruited at the University Hospital of Kiel. A detailed description of all BAV patients and controls analysed within this study is given as Supplementary material online, Cohort description.
2.2. Declaration
The authors state that their study complies with the Declaration of Helsinki, that the locally appointed ethics committees have approved all research protocols and that informed written consent has been obtained from all study participants before the inclusion of subjects into the study.
2.3. Genome-wide genotype data, quality control, and imputation
All patient samples (GWAS and replication) were genome-wide genotyped within this study. For the control cohorts (GWAS and replication) genotype data were obtained from previous studies. All genotyping arrays are summarized for each cohort in Supplementary material online, Methods.
The pre-imputation quality control (QC) is described in detail in Supplementary material online, Methods. After the QC, the sets of remaining SNPs were imputed using the TOPMed Imputation Server and TOPMed Reference panel17–19 for the German samples. For the UK samples, the Sanger Imputation Server and a combined reference panel using the Haplotype Reference Consortium release 1.1 (HRC r1.1)20 with additional variants from the UK10K and the 1000 Genomes phase 3 reference panel21 were used. After imputation, we excluded variants with a Hardy-Weinberg equilibrium deviation of P < 10−06 in patients or P < 10−04 in controls, a minor allele count < 20 or a minor allele frequency (MAF) in cases <0.01.
2.4. GWAS and GWAS meta-analysis
For the German cohort, trait associations were calculated with Firth logistic regression as implemented in Plink2,22 conditional on the first 10 variance standardized genotype principal components.
For the British cohort, logistic models were run in Plink222 utilizing Firth regression where models failed to converge. Analyses were adjusted for the first five genetic principle components.
The meta-analysis was performed with METAL23 using the fixed-effect inverse standard error-weighting approach, with a standard genome-wide significant threshold of 5 × 10−08.
To investigate whether independent associations are present in regions of genome-wide significance we carried out conditional analyses on the strongest associated SNP in each region with the meta-analysis approach COJO from GCTA.24,25 Here, we used the German cohort as a linkage disequilibrium (LD) reference sample. Additionally, we performed reciprocal conditional analysis with rs2293232 and rs2550262 on chromosome 3q29 as implemented in Plink2.22
2.5. LD score regression analyses
SNP-based heritability was calculated using LD score regression (LDSC).26 The reference files from GRCh37 were used as advised by the authors.
We next applied LDSC to partition the SNP-based heritability for BAV across transcriptome profiles from 16 foetal cardiac cell types that characterize heart development and were available through DESCARTES,27 the largest human single-cell atlas for foetal gene expression. Details of this analysis are provided as Supplementary material online, Methods.
2.6. FUMA analysis
We used FUMA (Functional Mapping and Annotation of Genome-Wide Association Studies)28,29 via the web-interface to gain functional insights into BAV. FUMA was used with default settings for the SNP2GENE and the GENE2FUNC modules. These tools provide information about functional annotations of associated SNPs and the possible biological role of the annotated genes.
2.7. Expression analysis at the BAV risk locus on chromosome 3q29 in embryonic/foetal and adult heart
Transcriptome data from single-cell RNA sequencing (scSeq) experiments derived from seven embryonic/foetal30 and two adult hearts31 were used. A detailed description of the tissue samples and scSeq protocols can be found in the corresponding publications.30,31 In brief, raw data of 676 single-cell transcriptomes isolated from seven human embryonic/foetal hearts (4.5–10 weeks of gestation) and three heart compartments (proximal outflow tract, ventricle, atria) were downloaded from the NCBI Sequence Read Archive (accession no. PRJNA510181). In addition, in-house data of 18 485 single-nuclei transcriptomes isolated from two adult human heart ventricles and atria were used. All transcriptomes were subjected to a standardized QC procedure followed by alignment to the Genome Reference Consortium Human Build 38 (hg38) using RNA STAR32 on the Galaxy web platform.33 This was followed by deduplication (BroadInstitue Picard tools: MarkDuplicates. https://broadinstitute.github.io/picard/Accessed August 15, 2020) and count matrix creation34 for single-cell data as well as the Cellranger pipeline35 with default settings including the –include-introns option for single-nuclei data, respectively. Seurat36 objects for all samples were created for downstream analyses from filtered count matrices. After quality filtering, the data were normalized and scaled and variable features for 669 embryonic/foetal and 16 896 adult single cells were detected using SCTransform.37 Data from embryonic and adult cardiac tissue were integrated using integration anchors and label transfer as described by Stuart et al.38 PCA and uniform manifold approximation and projection for dimension reduction were used to cluster cells into distinct biological identities and cell type identification was established based on the expression of known markers. For expression analysis of APOD, MUC20, MUC4, and TNK2, the ‘RNA’-assay of the integrated Seurat object was split into adult and embryonic cardiac cell populations retaining the clustering information of the ‘integrated’ -assay. The two populations were each normalized (NormalizeData) and the Seurat command FeaturePlot was used for visualization of gene expression with settings min.cutoff = ‘q10’ and max.cutoff = ‘q90’.
2.8. Zebrafish experiments
Handling of zebrafish was done in compliance with the guidelines from Directive 2010/63/EU of the European Parliament on the protection of animals used for scientific purposes and with German and Brandenburg state law, carefully monitored by the local authority for animal protection (LUGV, Brandenburg, Germany). Zebrafish embryos were euthanized by rapid chilling upon submersion into ice water bath (five parts ice/one part water, 2–4°C) for at least 20 min to ensure death. The Tg(kdrl:EGFP)s84339 zebrafish line was used and maintained under standard conditions as previously described.40 MOs (Gene Tools, LLC, USA) were diluted in ddH2O to 1 mM stock solution and injected into the yolk at the one-cell stage in 1 nL volume. Details of gRNAs, CRISPR/Cas9, and MOs sequences as well as experimental conditions, whole-mount immunohistochemistry, and whole-mount in situ hybridization protocol are provided in the Supplementary material online, Methods.
3. Results
3.1. GWAS single-marker association analysis
Our combined GWAS sample consisted of 2236 patients and 11 604 controls (see Methods for details). Genetic variants at three loci were associated with BAV at genome-wide significance. Two of the loci—on chromosome 1p21 and 8p23—have previously been reported,13,14 whereas the locus on chromosome 3q29 is novel. The findings for the lead variants at each locus are summarized in Table 1. The corresponding Q-Q- as well as Manhattan-plots are shown in Supplementary material online, Figures S1 and S2.
Table 1.
Lead associations of genome-wide significant and replicated BAV risk loci
| SNP | Chromosome [position in bp (hg38)] | Effect allele/other allele | Frequency in % | P-value | OR | 95% CI |
|---|---|---|---|---|---|---|
| rs11166276 | 1 (99 579 683) | T/C | 0.50 | 3.97 × 10−16 | 1.33 | 1.29–1.38 |
| rs13408842 | 2 (145 086 078) | A/G | 0.48 | 7.68 × 10−04 | 1.13 | 1.09–1.17 |
| rs2293232 | 3 (195 770 272) | T/C | 0.13 | 1.96 × 10−08 | 1.33 | 1.27–1.40 |
| rs2550262 | 3 (195 763 273) | C/A | 0.30 | 3.49 × 10−08 | 1.30 | 1.24–1.36 |
| rs6996406 | 8 (11 945 922) | A/G | 0.40 | 1.61 × 10−09 | 1.61 | 1.49–1.73 |
| rs3729856 | 8 (11 757 066) | G/A | 0.15 | 1.71 × 10−02 | 1.12 | 1.07–1.18 |
The associations are shown for the risk alleles (effect alleles). Mean allele frequencies for the risk alleles across all GWAS cohorts, P-values, odds ratios (OR), and the corresponding 95% confidence intervals (CIs) are shown.
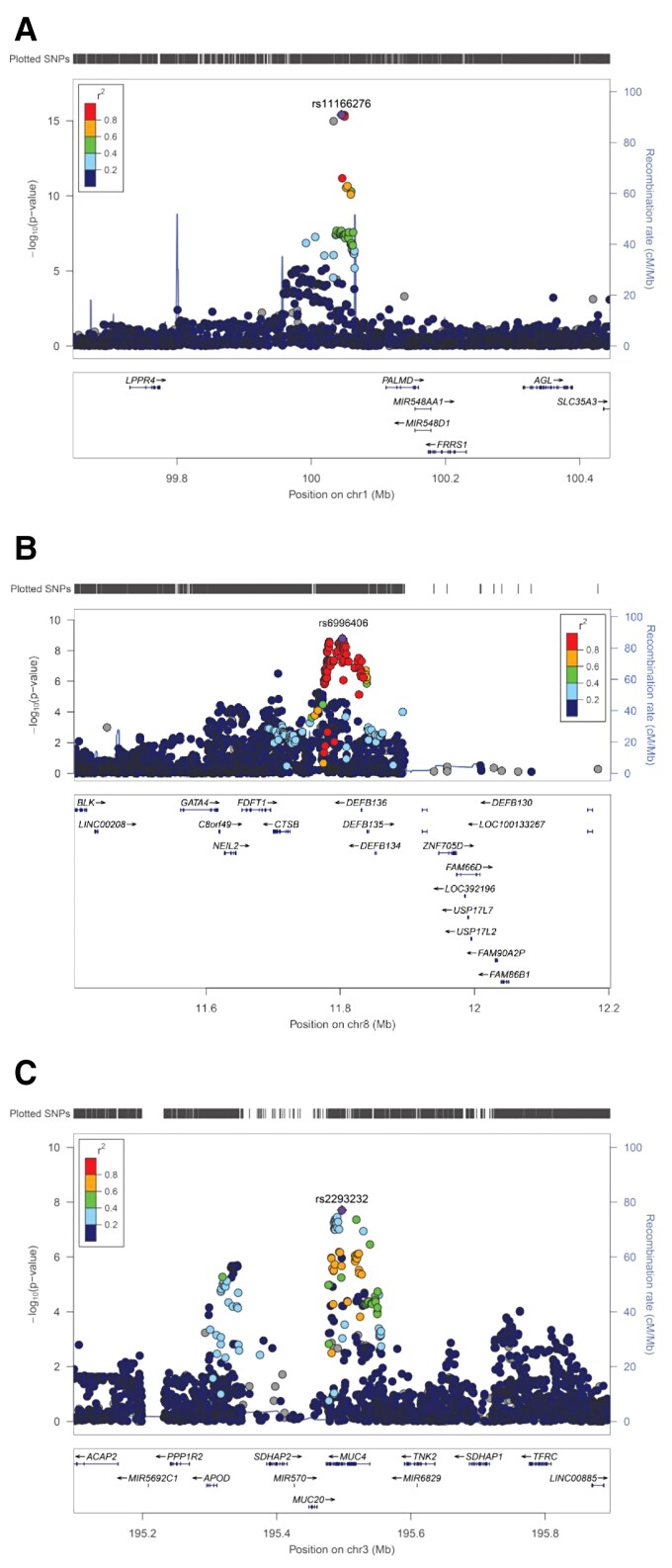
To assess the robustness of our findings, we first compared our data with previously published GWAS findings. SNP rs11166276 on chromosome 1p21 near PALMD showed the strongest genome-wide significant BAV association in our study [P = 3.97 × 10−16; odds ratio (OR) = 1.33] (Table 1 and Figure 1A). The risk variant is in perfect LD to rs7543130 (r2 = 1), which was the lead associated SNP in the GWAS of Helgadottir et al.14 On chromosome 8p23, rs6996406 showed genome-wide significant BAV association in our study (P = 1.61 × 10−09, OR = 1.61) (Table 1 and Figure 1B). The risk variant is in perfect LD to rs118065347 (r2 = 1), which was the lead SNP in the GWAS of Yang et al.13 and was linked to GATA4 as disease-contributing gene. Furthermore, we found nominal significant association at two additional BAV risk loci that have been reported previously.13,14 On chromosome 2q22, SNP rs13408842 within Intron 4 of TEX41, which was the lead variant at this locus in the GWAS of Helgadottir et al.,14 showed BAV association in our sample (P = 7.68 × 10−04, OR = 1.13) (Table 1). In addition, the missense variant rs3729856 (p.Ser377Gly) within Exon 6 of GATA4 on chromosome 8p23 that showed independent disease association at this locus in the GWAS of Yang et al.13 was significant BAV associated in our sample (P = 1.71 × 10−02, OR = 1.12) (Table 1). Thus, all common risk loci at PALMD, TEX41, and GATA413 that have been previously been found in BAV GWAS were replicated in our data set. However, we could not test the previously described risk SNPs rs137867582 (p.Thr1221Met) within DHX3813 and rs387906656 (p.Arg721Trp) within MYH6,14 because the MAFs of the corresponding risk alleles are extremely rare in the European population (<0.001% according to gnomAD41).
Figure 1.
Regional association plots of BAV risk loci on chromosome 1p21 near PALMD (A), 8p23 near GATA4 (B) and 3q29 within MUC4 (C). At each locus, associations [−log10(P-values)] are shown for SNPs flanking 400 kb on either side of the lead associated SNP. The lead variant is indicated by the corresponding rs-number. Other markers at each locus are displayed by different colours, which indicate different levels of LD (r2) to the lead SNP. Furthermore, all annotated genes within each region are shown with arrows indicating their transcription direction.
At the novel risk locus on chromosome 3q29 rs2293232 showed genome-wide significant BAV association (P = 1.96 × 10−08, OR = 1.33) (Table 1 and Figure 1C). This SNP is located in Exon 5 of MUC4 and represents a missense variant (p.Ala4448Thr) that is predicted to be benign according to gnomAD.41 In addition, a second and independent variant at this locus showed genome-wide significant BAV association (P = 3.49 × 10−08, OR = 1.30) (Table 1). This SNP, rs2550262, is only in weak LD to rs2293232 (r2 = 0.53). Accordingly, both variants showed significant BAV association upon reciprocal conditioning (Prs2293232 = 4.96 × 10−02, Prs2550262 = 1.12 × 10−03). Of note, rs2550262 is in nearly perfect LD to rs2246901 (r2 = 0.97), a missense variant in Exon 13 of MUC4 (p.Ala4821Ser) and deleterious according to gnomAD.41 Thus, this variant represents the most likely causal SNP at this locus.
Finally, we analysed 421 independent European BAV patients and 3643 ethnically matched controls in order to replicate the newly identified risk locus on chromosome 3q29. Here, we found independent BAV association for rs2550262 (P = 1.36 × 10−02, OR = 1.22), one of the lead GWAS SNPs within MUC4. In addition, rs2246901, which is in nearly perfect LD to rs2550262 and represents the most likely causal SNP at this locus, showed significant BAV association (P = 1.31 × 10−02, OR = 1.22). In the combined sample (GWAS and replication) rs2550262 was BAV associated with P = 1.85 × 10−09 (OR = 1.28) (see Supplementary material online, Table S2).
3.2. Genetic BAV architecture on the polygenic level
We used the LDSC method, which allows the collective analysis of common GWAS variants, to estimate the SNP-based heritability of BAV and to test whether GWAS risk SNPs are enriched in regions surrounding genes with specific expression in different cell types of the foetal heart.
Based on our genome-wide data, we estimate the SNP-based heritability of BAV to be 9.55 ± 3.97% standard deviation (SD). The reported BAV heritability, which is based on family studies, ranges from 47 to 89%4,5 with the lower estimate referring to complex genetic BAV forms, while the upper estimate reflects monogenic forms of BAV. Thus, our data already explain 20% of the reported heritability (h2 = 47%).
Next, we used DESCARTES27 to assess cell type-specific GWAS enrichment. This database provide an annotation of cell type-specific transcriptome profiles for 15 organs from 121 foetal tissues. Within the foetal heart, DESCARTES identified 16 cardiac cell types with distinct transcriptome profiles during development. We applied LDSC to partition the SNP-based heritability for BAV across these 16 transcriptome profiles and found nominally significant GWAS enrichment (P < 0.05) among 11 foetal cardiac cell types (see Supplementary material online, Table S3). Of note, we observed only a weak overlap of expressed genes between these cell types (see Supplementary material online, Figure S3). Of the implicated cell types, the transcriptome profile in vascular endothelial cells also showed significant enrichment for BAV GWAS signals after a Bonferroni-correction for multiple testing (Puncorrected = 5.9 × 10−04, Pcorrected = 9.5 × 10−03).
In addition to the LDSC method, we analysed the GWAS data on the polygenic level using FUMA,28,29 a tool that uses functional and biological information as well as GWAS data to prioritize disease genes. The FUMA gene-based analysis was performed for a total of 18 830 genes. Here, two genes were implicated after correction for multiple testing (P < 0.05/18 830 or P < 2.66 × 10−06) (see Supplementary material online, Figure S4). One refers to MUC4, the BAV candidate gene on chromosome 3q29 identified by the GWAS, where FUMA showed significant BAV association with P = 9.66 × 10−08. The other gene that showed significant BAV association in FUMA is DCAF4 on chromosome 14q24 (P = 1.54 × 10−06). In contrast, all FUMA tissue expression profile analyses did not reveal any significant or biologically interesting results (data not shown), which could be because FUMA uses information only from adult heart tissue.
3.3. Expression of genes at the BAV risk locus on chromosome 3q29 in embryonic/foetal and adult heart
We used transcriptome data from scSeq experiments derived from seven embryonic/foetal and two adult hearts30,31 of which expression data were available. Supplementary material online, Figure S5A shows that the cell type-specific embryonic/foetal and adult cardiac transcriptome data lead to perfectly superimposed clusters, which have been published previously.31 We then focused on expression data of genes from the embryonic/foetal and adult heart, in which variants showed at least weak LD (r2 > 0.2) to the strongest BAV-associated SNP on chromosome 3q29. According to Supplementary material online, Figure 1B, these are the genes APOD, SDHAP2, MUC20, MUC4, and TNK2. Of them, all—except SDHAP2—showed expression in embryonic/foetal and adult cardiac cell types, while MUC4 represented the gene that—compared with the other genes—was expressed in embryonic/foetal cardiac cells but almost undetectable in the adult heart (see Supplementary material online, Figure S5C). Thus, also based on these expression data MUC4 represents an interesting BAV candidate gene at the newly identified GWAS locus, although the other genes cannot be excluded as the true risk conferring BAV genes.
3.4. Cardiac valvulogenesis is delayed upon loss of Muc4 in zebrafish
In the GWAS and replication study, SNP rs2550262 in MUC4 was significantly BAV associated and in strong LD to the missense variant rs2246901 (p.Ala4821Ser) that is deleterious according to gnomAD.41 In addition, MUC4 showed almost specific expression in embryonic/foetal cardiac cells compared with adult heart tissue. We, thus, selected MUC4 for first functional testing of its potential involvement on valve leaflet morphogenesis in zebrafish.
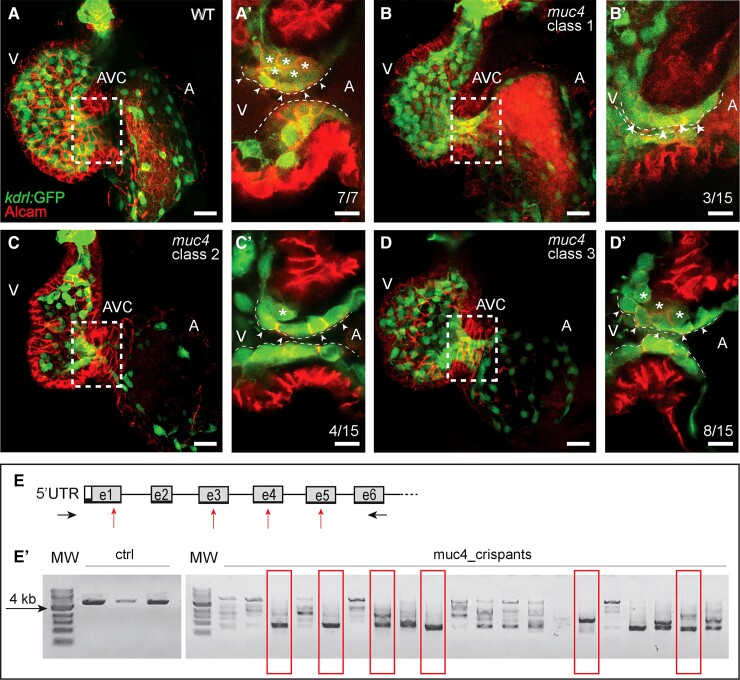
Previous transcriptome data on cardiac tissue revealed that the predicted zebrafish orthologous gene si:ch73-105b23.6 (here called muc4) is expressed at 21.5, 54, and 72 h post-fertilization (hpf) during stages of atrioventricular (AV) valvulogenesis.42–44 To functionally study the role of Muc4 during valvulogenesis, we used two different loss-of-function strategies. First, we used a CRISPR/Cas9-mediated knockout (Figure 2) that was based on injecting of a mixed pool of four gRNAs and Cas9 protein. We tested the efficacy of the CRISPR/Cas9-mediated gene knockout on a total of 58 embryos. Genomic PCR of the muc4 locus revealed that the mixture of four gRNAs had induced mutations in almost all embryos. Among these, we chose 15 embryos that had not only a large deletion at the muc4 locus but also lacked a WT allele (Figure 2E and E’). When we characterized these 15 muc4 crispant embryos, we found that the gene knockout had not caused any gross defects in cardiac morphogenesis at 58 hpf. However, careful analysis of endocardial cell ingression into the cardiac jelly during the morphogenesis of the AV valve leaflets revealed a delay in muc4 crispants compared with wild-type (WT) non-injected control embryos (Figure 2A–D). Whereas in WT endocardial cell ingression has produced an abluminal layer of endocardial cells at that stage (Figure 2A’, asterisks), muc4 crispants exhibited a delayed endocardial cell ingression into the cardiac jelly. We identified three phenotype classes among the 15 muc4 crispants as quantified in Figure 2B and D. As an alternative approach, we also designed three non-overlapping MO antisense oligonucleotides to target muc4 pre-mRNA splice sites (see Supplementary material online, Figure S6). For this alternative MO knockdown approach (see Supplementary material online, Figure S6), a similar delay in endocardial cell ingression during AV valvulogenesis was observed (as quantified in see Supplementary material online, Figure S7). Thus, the muc4 crispant and morphant AV valvular phenotypes correlate with a temporal delay in cardiac valve development.
Figure 2.
Zebrafish muc4 knockout using CRISPR/Cas9. (A–D) Maximum intensity projections from confocal z-stack images of hearts at 58 hpf. (A’–D’) Single confocal z-section images of the AVC region. Endocardial cells in luminal positions (arrowheads) at the AVC are marked by kdrl:GFP and Alcam.45 Endocardial cells that have already invaded the extracellular matrix and are in abluminal positions are marked with asterisks. (A’) While in WT control embryos many endocardial cells are in abluminal positions, some of the 15 muc4 crispants with a complete truncation of the genomic locus exhibit a delayed ingression of endocardial cells into the cardiac jelly. The severity of the phenotypes varied according to the following classes: (B’) Class 1 embryos (3 of 15) lack an endocardial cell ingression by that stage; (C’) Class 2 embryos (4 of 15), have only a single endocardial cell ingressing into the cardiac jelly at that stage; (D’) in muc4 crispants of class 3 (8 of 15), a few endocardial cells have ingressed into the cardiac jelly. V, ventricle; A, atrium; AVC, atrioventricular canal. Scale bars are 20 µm (A–D) and 10 µm (A’–D’). (E) Schematic representation of the zebrafish muc4 locus and gRNA binding sites (red arrows). Black arrows indicate the positions of primers used to assess the efficacy of CRISPS-induced knockout by PCR. (E’) Single embryo PCR products from control embryos (ctrl) and embryos injected with a mixture of four gRNAs and Cas9 protein (muc4_crispants). Red boxes are indicating samples that lack any WT genomic amplification bands. Among 58 injected embryos that were genotyped, 15 had a large truncation of the genomic region and did not contain any WT band. Only these crispants were used for phenotypic characterization. MW, molecular weight.
4. Discussion
Using the largest GWAS sample analysed so far, we independently confirmed all common loci that have previously been reported to contribute to the risk for BAV disease. In addition, we identified a novel risk locus on chromosome 3q29 and revealed that MUC4 at this locus might be involved in embryonic valvulogenesis. Finally, our results underline the polygenic nature in BAV development.
Although recent GWAS have identified risk variants for BAV development near PALMD (chromosome 1p21) and TEX41 (chromosome 2q22), functional data are still lacking to determine the causal BAV genes at both loci. Helgadottir et al.14 used chromatin interaction maps from different foetal and adult heart tissues and found that the BAV risk variants at both loci are located within regulatory regions that interact with promoters on neighbouring genes. On chromosome 1p21 this involved promoters of PALMD, SNX7, PLPPR5, and PLPPR4 as well as two non-coding RNAs. On chromosome 2q22 promoters of ZEB2 and GTDC1 as well as three non-coding RNAs, including TEX41 have been implicated. In contrast, GATA4 on chromosome 8p23 is so far the only gene at a GWAS locus with convincing evidence of being involved in BAV development. Common risk variants and rare deleterious mutations in GATA4 have been repeatedly found in various CHDs, including BAV, atrial and ventricular septal defect as well as Tetralogy of Fallot.46–51
In the present study, we identified and independently confirmed a fourth BAV risk locus on chromosome 3q29. Based on our GWAS data, expression data from embryonic/foetal and adult heart tissues as well as zebrafish knockout and knockdown experiments, MUC4 represents an interesting BAV candidate gene at this locus. SNP rs2246901 in Exon 13 of MUC4 is in nearly perfect LD to the lead-associated GWAS variant and represents a missense variant (p.Ala4821Ser) that is deleterious according to gnomAD.41 In line with this finding, knockout and knockdown of this gene in zebrafish revealed that muc4 may impact cardiac AV valve leaflet morphogenesis causing a developmental delay of the multi-layering of valve leaflets. The zebrafish heart is two-chambered containing three sets of valves (AV, inflow, and outflow tract valves). In our study, we focused on the development of the AV valve between atrial and ventricular chambers because the development of this valve has been well characterized and it forms at an early developmental stage between 48 and 72 hpf. Of note, zebrafish outflow tract valve leaflets, which are related to human aortic valves affected in BAV, emerge and elongate between 96 and 144 hpf and, currently much less is known about their mode of development.52 We also found that by this stage that AV valve development had partially recovered in our Muc4 knockout and knockdown zebrafish models. On the cellular level, MUC4 also represents an interesting BAV candidate gene. MUC4 is involved in epithelial-to-mesenchymal transformation (EMT),53,54 a cellular process that is of major relevance for cardiac valve development.55–57 During EMT, the molecular phenotype of the endocardial epithelial cells changes towards a mesenchymal character and those cells migrate from the inner lining of the endocardium towards the cardiac jelly forming the so-called endocardial cushions.58 At later stages fusion of these cardiac cushions results in the formation of valve leaflets.58 Interestingly, induced molecular changes in the composition of the prevalvular mesenchyme can result in the formation of BAVs.59 A similar transition of cells in adult life can be seen during cancer and especially in tumour metastasis. In this context expression of MUC4 has been previously shown to initiate the process of EMT in cancer cells,53 whereas inhibition of MUC4 expression resulted in suppressed cell invasion and EMT.54
Although our genetic and zebrafish data indicate MUC4 as an interesting BAV candidate gene, Muc4 knockout mice are viable, fertile, and show no obvious developmental abnormalities.60,61 However, mild cardiac valve abnormalities may have been missed in the latter studies. In muc4 knockout and knockdown zebrafish models, the developmental phenotype only became apparent after focused cardiac phenotyping.
In addition, our study represents the first that examined the genetic BAV architecture beyond the single-marker level. Our findings show that a substantial fraction of BAV heritability is polygenic with an estimated SNP-based heritability of 9.55 ± 3.97% (SD) that already explains around 20% of the observed BAV heritability. In addition, we found that the transcriptome profile of foetal endothelial cells in DESCARTES27 showed significant enrichment of GWAS signals. This implies that endothelial cells, which along with interstitial cells are the most common aortic valve cell type,62 are of major relevance for BAV development. However, the transcriptome profiles of 10 additional foetal heart cell types showed nominally significant enrichment of BAV GWAS signals. This might point to a complex interaction of different cell types involved in BAV development and might also explain why BAV patients often present with other CHDs, where additional disease-relevant cell types are involved.
Our study has two main limitations. Most of our patients were symptomatic cases, mainly with an AS phenotype. Even though the vast majority of BAV patients will become symptomatic, genetic variability also influences the BAV outcome. We, thus, cannot exclude the possibility that we have also identified genetic risk factors that influence the occurrence of BAV complications rather than its development. In addition, our study did not allow the identification of rare genetic risk factors contributing to BAV. We, thus, could not examine the contribution of the previously described rare risk variants in DHX38 and MYH6. Large exome-wide sequencing studies will provide corresponding data in the future.
5. Conclusions
We independently confirm that the risk loci at PALMD, TEX41, and GATA4 contribute to BAV development. In addition, our results imply that a disturbed MUC4 function contributes to BAV development. The BAV-associated missense variant rs2246901 (p.Ala4821Ser) in MUC4 is predicted to be deleterious and Muc4 knockout as well as knockdown in zebrafish led to a persistent delay in AV valvular development. However, further studies are needed to determine the relevance of MUC4 in BAV development. The present study constitutes the basis for these future studies. Furthermore, we show that a substantial fraction of BAV heritability is polygenic and around 20% of the observed heritability can be explained by our GWAS data. In addition, our findings point to the transcriptome profile in foetal endothelial cells as a major source contributing to BAV pathology.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Authors’ contribution
Design of the work, acquisition/analysis/interpretation of data for the work, drafting the manuscript, final approval of the manuscript, being accountable for the parts of the work she/he has done: J.G., A.S., R.D., F.F., D.S., M.K., J.-M.S., G.N., M.M.N., A.P.B., S.A.-S., N.J.S., J.E., T.T., J.S. Acquisition of data/study participants for the work, final approval of the manuscript, being accountable for the parts of the work she/he has done: C.P.N., B.A.-K., A.-S.G., C.M.H.B., C.F.Z., L.L.K., P.S.B., T.R.W., S.H., S.E., B.F., S.A.M., M.S., H.K., M.S., F.A.K., P.N., L.B., M.K., V.V., M.A., S.B., K.-L.L., Y.H., M.K., U.M., L.O.C., I.B., C.L., P.U., W.-K.K., J.K., D.E., U.N.-G., P.H., F.W., S.D., H.L., M.D., M.v.S., K.K., T.K., C.H., H.S.
Supplementary Material
Acknowledgements
The authors thank all study participants who contributed to this study.
Contributor Information
Jan Gehlen, Institute of Human Genetics, University of Bonn and University Hospital Bonn, Bonn, Germany; Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Anja Stundl, Department of Medicine II, Heart Center Bonn, University of Bonn and University Hospital Bonn, Bonn, Germany; Klinik und Poliklinik für Innere Medizin I, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany; DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany.
Radoslaw Debiec, Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK; East Midlands Congenital Heart Centre, Glenfield Hospital, Leicester, UK.
Federica Fontana, Institute of Biochemistry and Biology, Potsdam University, Potsdam, Germany.
Markus Krane, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Division of Experimental Surgery, Department of Cardiovascular Surgery, Institute Insure, German Heart Center Munich, TUM School of Medicine, Technical University of Munich, Munich, Germany; Division of Cardiac Surgery, Department of Surgery, Yale University School of Medicine, New Haven, CT, USA.
Dinara Sharipova, Institute of Biochemistry and Biology, Potsdam University, Potsdam, Germany.
Christopher P Nelson, Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
Baravan Al-Kassou, Department of Medicine II, Heart Center Bonn, University of Bonn and University Hospital Bonn, Bonn, Germany.
Ann-Sophie Giel, Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Jan-Malte Sinning, Department of Medicine II, Heart Center Bonn, University of Bonn and University Hospital Bonn, Bonn, Germany.
Christopher M H Bruenger, Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Carolin F Zelck, Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Laura L Koebbe, Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Peter S Braund, Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
Thomas R Webb, Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
Simon Hetherington, Kettering General Hospital NHS Foundation Trust, Kettering, UK.
Stephan Ensminger, Department of Cardiac and Thoracic Vascular Surgery, University Heart Center Lübeck, University Hospital of Schleswig-Holstein, Lübeck, Germany; DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Lübeck, Germany.
Buntaro Fujita, Department of Cardiac and Thoracic Vascular Surgery, University Heart Center Lübeck, University Hospital of Schleswig-Holstein, Lübeck, Germany; DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Lübeck, Germany.
Salah A Mohamed, Department of Cardiac and Thoracic Vascular Surgery, University Heart Center Lübeck, University Hospital of Schleswig-Holstein, Lübeck, Germany; DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Lübeck, Germany.
Malakh Shrestha, Department of Adult and Pediatric Cardiothoracic Surgery, Vascular Surgery, Heart and Lung Transplantation, Hannover Medical School, Hannover, Germany.
Heike Krueger, Department of Adult and Pediatric Cardiothoracic Surgery, Vascular Surgery, Heart and Lung Transplantation, Hannover Medical School, Hannover, Germany.
Matthias Siepe, Heart Center Freiburg/Bad Krozingen, University Freiburg/Bad Krozingen, Freiburg, Germany.
Fabian Alexander Kari, Heart Center Freiburg/Bad Krozingen, University Freiburg/Bad Krozingen, Freiburg, Germany.
Peter Nordbeck, Medizinische Klinik und Poliklinik I, University Hospital Würzburg, Würzburg, Germany.
Larissa Buravezky, Medizinische Klinik und Poliklinik I, University Hospital Würzburg, Würzburg, Germany.
Malte Kelm, Department of Cardiology, Pneumology and Angiology, University Hospital Duesseldorf, Duesseldorf, Germany.
Verena Veulemans, Department of Cardiology, Pneumology and Angiology, University Hospital Duesseldorf, Duesseldorf, Germany.
Matti Adam, Department of Medicine III, Heart Center Cologne, University Hospital Cologne, Cologne, Germany.
Stephan Baldus, Department of Medicine III, Heart Center Cologne, University Hospital Cologne, Cologne, Germany.
Karl-Ludwig Laugwitz, Klinik und Poliklinik für Innere Medizin I, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany; DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany.
Yannick Haas, Klinik und Poliklinik für Innere Medizin I, Klinikum rechts der Isar, Technical University of Munich, Munich, Germany.
Matthias Karck, Department of Cardiothoracic Surgery, University Hospital Heidelberg, Heidelberg, Germany.
Uwe Mehlhorn, Department of Cardiothoracic Surgery, Helios Klinik Karlsruhe, Karlsruhe, Germany.
Lars Oliver Conzelmann, Department of Cardiothoracic Surgery, Helios Klinik Karlsruhe, Karlsruhe, Germany.
Ingo Breitenbach, Department of Cardiothoracic Surgery and Vascular Surgery, Clinic of Braunschweig, Braunschweig, Germany.
Corinna Lebherz, Department of Medicine I, Cardiology/Angiology/Intensive Care, University Hospital Aachen, Aachen, Germany.
Paul Urbanski, Department of Cardiovascular Surgery, Cardiovascular Clinic, Rhön-Klinikum Campus Bad Neustadt, Neustadt, Germany.
Won-Keun Kim, Department of Cardiology, Heart Center, Kerckhoff Clinic, Bad Nauheim, Germany.
Joscha Kandels, Department of Cardiology, University Hospital Leipzig, Leipzig, Germany.
David Ellinghaus, Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Kiel, Germany; Novo Nordisk Foundation Center for Protein Research, Disease Systems Biology, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark.
Ulrike Nowak-Goettl, Department of Clinical Chemistry, Thrombosis and Hemostasis Unit, University Hospital of Kiel and Lübeck, Kiel, Germany.
Per Hoffmann, Institute of Human Genetics, University of Bonn and University Hospital Bonn, Bonn, Germany.
Felix Wirth, Division of Experimental Surgery, Department of Cardiovascular Surgery, Institute Insure, German Heart Center Munich, TUM School of Medicine, Technical University of Munich, Munich, Germany.
Stefanie Doppler, Division of Experimental Surgery, Department of Cardiovascular Surgery, Institute Insure, German Heart Center Munich, TUM School of Medicine, Technical University of Munich, Munich, Germany.
Harald Lahm, Division of Experimental Surgery, Department of Cardiovascular Surgery, Institute Insure, German Heart Center Munich, TUM School of Medicine, Technical University of Munich, Munich, Germany.
Martina Dreßen, Division of Experimental Surgery, Department of Cardiovascular Surgery, Institute Insure, German Heart Center Munich, TUM School of Medicine, Technical University of Munich, Munich, Germany.
Moritz von Scheidt, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Department of Cardiology, German Heart Centre Munich, Technical University of Munich, Munich, Germany.
Katharina Knoll, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Department of Cardiology, German Heart Centre Munich, Technical University of Munich, Munich, Germany.
Thorsten Kessler, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Department of Cardiology, German Heart Centre Munich, Technical University of Munich, Munich, Germany.
Christian Hengstenberg, Division of Cardiology, Department of Internal Medicine II, Medical University of Vienna, Vienna, Austria.
Heribert Schunkert, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Department of Cardiology, German Heart Centre Munich, Technical University of Munich, Munich, Germany.
Georg Nickenig, Department of Medicine II, Heart Center Bonn, University of Bonn and University Hospital Bonn, Bonn, Germany.
Markus M Nöthen, Institute of Human Genetics, University of Bonn and University Hospital Bonn, Bonn, Germany.
Aidan P Bolger, NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK; East Midlands Congenital Heart Centre, Glenfield Hospital, Leicester, UK; Institute of Biochemistry and Biology, Potsdam University, Potsdam, Germany.
Salim Abdelilah-Seyfried, Institute of Biochemistry and Biology, Potsdam University, Potsdam, Germany; Institute of Molecular Biology, Hannover Medical School, Hannover, Germany.
Nilesh J Samani, Department of Cardiovascular Sciences, University of Leicester, Leicester, UK; NIHR Leicester Biomedical Research Centre, Glenfield Hospital, Leicester, UK.
Jeanette Erdmann, DZHK (German Centre for Cardiovascular Research), Partner Site Hamburg/Kiel/Lübeck, Lübeck, Germany; Institute for Cardiogenetics, University Heart Centre Lübeck, University of Lübeck, Lübeck, Germany.
Teresa Trenkwalder, DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, Munich, Germany; Department of Cardiology, German Heart Centre Munich, Technical University of Munich, Munich, Germany.
Johannes Schumacher, Institute of Human Genetics, University of Bonn and University Hospital Bonn, Bonn, Germany; Institute of Human Genetics, Philipps University of Marburg, Marburg, Germany.
Funding
This work was supported by the following grants from the Deutsche Forschungsgemeinschaft (DFG): TRR259, SFB958, SE2016/13-1, SE2016/17-1, NO246/17-1, KR3985/12-1, EXC 2167-390884018, INST 336/104-1, and INST 336/114-1 FUGG. The Leicester samples were collected as part of the BRAVE study and supported by the NIHR Leicester Biomedical Research Centre. R.D. was supported by the Leicester Biomedical Research Centre. C.P.N. and T.R.W. are funded by the British Heart Foundation.
Data availability
The data that support the findings of this study are available on request from the corresponding author (J.S.).
References
- 1. Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol 2010;55:2789–2800. [DOI] [PubMed] [Google Scholar]
- 2. Ward C. Clinical significance of the bicuspid aortic valve. Heart 2000;83:81–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roberts WC, Ko JM. Frequency by decades of unicuspid, bicuspid, and tricuspid aortic valves in adults having isolated aortic valve replacement for aortic stenosis, with or without associated aortic regurgitation. Circulation 2005;111:920–925. [DOI] [PubMed] [Google Scholar]
- 4. Cripe L, Andelfinger G, Martin LJ, Shooner K, Benson DW. Bicuspid aortic valve is heritable. J Am Coll Cardiol 2004;44:138–143. [DOI] [PubMed] [Google Scholar]
- 5. Galian-Gay L, Carro Hevia A, Teixido-Tura G, Rodriguez Palomares J, Gutierrez-Moreno L, Maldonado G, Gonzalez-Alujas MT, Sao-Aviles A, Gallego P, Calvo-Iglesias F, Bermejo J, Robledo-Carmona J, Sanchez V, Saura D, Sevilla T, Burillo-Sanz S, Guala A, Garcia-Dorado D, Evangelista A, BICUSPID investigators . Familial clustering of bicuspid aortic valve and its relationship with aortic dilation in first-degree relatives. Heart 2019;105:603–608. [DOI] [PubMed] [Google Scholar]
- 6. Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, Bourgeois S, Estrera AL, Safi HJ, Sparks E, Amor D, Ades L, McConnell V, Willoughby CE, Abuelo D, Willing M, Lewis RA, Kim DH, Scherer S, Tung PP, Ahn C, Buja LM, Raman CS, Shete SS, Milewicz DM. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet 2007;39:1488–1493. [DOI] [PubMed] [Google Scholar]
- 7. Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, Xu FL, Myers LA, Spevak PJ, Cameron DE, De Backer J, Hellemans J, Chen Y, Davis EC, Webb CL, Kress W, Coucke P, Rifkin DB, De Paepe AM, Dietz HC. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet 2005;37:275–281. [DOI] [PubMed] [Google Scholar]
- 8. Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature 2005;437:270–274. [DOI] [PubMed] [Google Scholar]
- 9. Gould RA, Aziz H, Woods CE, Seman-Senderos MA, Sparks E, Preuss C, Wunnemann F, Bedja D, Moats CR, McClymont SA, Rose R, Sobreira N, Ling H, MacCarrick G, Kumar AA, Luyckx I, Cannaerts E, Verstraeten A, Bjork HM, Lehsau AC, Jaskula-Ranga V, Lauridsen H, Shah AA, Bennett CL, Ellinor PT, Lin H, Isselbacher EM, Lino Cardenas CL, Butcher JT, Hughes GC, Lindsay ME, Baylor-Hopkins Center for Mendelian Genomics, MIBAVA Leducq Consortium, Mertens L, Franco-Cereceda A, Verhagen JMA, Wessels M, Mohamed SA, Eriksson P, Mital S, Van Laer L, Loeys BL, Andelfinger G, McCallion AS, Dietz HC. ROBO4 variants predispose individuals to bicuspid aortic valve and thoracic aortic aneurysm. Nat Genet 2019; 51:42–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tan HL, Glen E, Topf A, Hall D, O’Sullivan JJ, Sneddon L, Wren C, Avery P, Lewis RJ, ten Dijke P, Arthur HM, Goodship JA, Keavney BD. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat 2012; 33:720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wunnemann F, Ta-Shma A, Preuss C, Leclerc S, van Vliet PP, Oneglia A, Thibeault M, Nordquist E, Lincoln J, Scharfenberg F, Becker-Pauly C, Hofmann P, Hoff K, Audain E, Kramer HH, Makalowski W, Nir A, Gerety SS, Hurles M, Comes J, Fournier A, Osinska H, Robins J, Puceat M, MIBAVA Leducq Consortium principal investigators, Elpeleg O, Hitz MP, Andelfinger G. Loss of ADAMTS19 causes progressive non-syndromic heart valve disease. Nat Genet 2020; 52:40–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prakash SK, Bosse Y, Muehlschlegel JD, Michelena HI, Limongelli G, Della Corte A, Pluchinotta FR, Russo MG, Evangelista A, Benson DW, Body SC, Milewicz DM, Investigators BA. A roadmap to investigate the genetic basis of bicuspid aortic valve and its complications: insights from the International BAVCon (Bicuspid Aortic Valve Consortium). J Am Coll Cardiol 2014;64:832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang B, Zhou W, Jiao J, Nielsen JB, Mathis MR, Heydarpour M, Lettre G, Folkersen L, Prakash S, Schurmann C, Fritsche L, Farnum GA, Lin M, Othman M, Hornsby W, Driscoll A, Levasseur A, Thomas M, Farhat L, Dube MP, Isselbacher EM, Franco-Cereceda A, Guo DC, Bottinger EP, Deeb GM, Booher A, Kheterpal S, Chen YE, Kang HM, Kitzman J, Cordell HJ, Keavney BD, Goodship JA, Ganesh SK, Abecasis G, Eagle KA, Boyle AP, Loos RJF, Eriksson P, Tardif JC, Brummett CM, Milewicz DM, Body SC, Willer CJ. Protein-altering and regulatory genetic variants near GATA4 implicated in bicuspid aortic valve. Nat Commun 2017;8:15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Helgadottir A, Thorleifsson G, Gretarsdottir S, Stefansson OA, Tragante V, Thorolfsdottir RB, Jonsdottir I, Bjornsson T, Steinthorsdottir V, Verweij N, Nielsen JB, Zhou W, Folkersen L, Martinsson A, Heydarpour M, Prakash S, Oskarsson G, Gudbjartsson T, Geirsson A, Olafsson I, Sigurdsson EL, Almgren P, Melander O, Franco-Cereceda A, Hamsten A, Fritsche L, Lin M, Yang B, Hornsby W, Guo D, Brummett CM, Abecasis G, Mathis M, Milewicz D, Body SC, Eriksson P, Willer CJ, Hveem K, Newton-Cheh C, Smith JG, Danielsen R, Thorgeirsson G, Thorsteinsdottir U, Gudbjartsson DF, Holm H, Stefansson K. Genome-wide analysis yields new loci associating with aortic valve stenosis. Nat Commun 2018;9:987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmermund A, Mohlenkamp S, Stang A, Gronemeyer D, Seibel R, Hirche H, Mann K, Siffert W, Lauterbach K, Siegrist J, Jockel KH, Erbel R. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL study. Risk factors, evaluation of coronary calcium and lifestyle. Am Heart J 2002;144:212–218. [DOI] [PubMed] [Google Scholar]
- 16. Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, Schlessinger D, Stambolian D, Loh PR, Iacono WG, Swaroop A, Scott LJ, Cucca F, Kronenberg F, Boehnke M, Abecasis GR, Fuchsberger C. Next-generation genotype imputation service and methods. Nat Genet 2016;48:1284–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taliun D, Harris DN, Kessler MD, Carlson J, Szpiech ZA, Torres R, Taliun SAG, Corvelo A, Gogarten SM, Kang HM, Pitsillides AN, LeFaive J, Lee SB, Tian X, Browning BL, Das S, Emde AK, Clarke WE, Loesch DP, Shetty AC, Blackwell TW, Smith AV, Wong Q, Liu X, Conomos MP, Bobo DM, Aguet F, Albert C, Alonso A, Ardlie KG, Arking DE, Aslibekyan S, Auer PL, Barnard J, Barr RG, Barwick L, Becker LC, Beer RL, Benjamin EJ, Bielak LF, Blangero J, Boehnke M, Bowden DW, Brody JA, Burchard EG, Cade BE, Casella JF, Chalazan B, Chasman DI, Chen YI, Cho MH, Choi SH, Chung MK, Clish CB, Correa A, Curran JE, Custer B, Darbar D, Daya M, de Andrade M, DeMeo DL, Dutcher SK, Ellinor PT, Emery LS, Eng C, Fatkin D, Fingerlin T, Forer L, Fornage M, Franceschini N, Fuchsberger C, Fullerton SM, Germer S, Gladwin MT, Gottlieb DJ, Guo X, Hall ME, He J, Heard-Costa NL, Heckbert SR, Irvin MR, Johnsen JM, Johnson AD, Kaplan R, Kardia SLR, Kelly T, Kelly S, Kenny EE, Kiel DP, Klemmer R, Konkle BA, Kooperberg C, Kottgen A, Lange LA, Lasky-Su J, Levy D, Lin X, Lin KH, Liu C, Loos RJF, Garman L, Gerszten R, Lubitz SA, Lunetta KL, Mak ACY, Manichaikul A, Manning AK, Mathias RA, McManus DD, McGarvey ST, Meigs JB, Meyers DA, Mikulla JL, Minear MA, Mitchell BD, Mohanty S, Montasser ME, Montgomery C, Morrison AC, Murabito JM, Natale A, Natarajan P, Nelson SC, North KE, O’Connell JR, Palmer ND, Pankratz N, Peloso GM, Peyser PA, Pleiness J, Post WS, Psaty BM, Rao DC, Redline S, Reiner AP, Roden D, Rotter JI, Ruczinski I, Sarnowski C, Schoenherr S, Schwartz DA, Seo JS, Seshadri S, Sheehan VA, Sheu WH, Shoemaker MB, Smith NL, Smith JA, Sotoodehnia N, Stilp AM, Tang W, Taylor KD, Telen M, Thornton TA, Tracy RP, Van Den Berg DJ, Vasan RS, Viaud-Martinez KA, Vrieze S, Weeks DE, Weir BS, Weiss ST, Weng LC, Willer CJ, Zhang Y, Zhao X, Arnett DK, Ashley-Koch AE, Barnes KC, Boerwinkle E, Gabriel S, Gibbs R, Rice KM, Rich SS, Silverman EK, Qasba P, Gan W, NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium; Papanicolaou GJ, Nickerson DA, Browning SR, Zody MC, Zollner S, Wilson JG, Cupples LA, Laurie CC, Jaquish CE, Hernandez RD, O’Connor TD, Abecasis GR. Sequencing of;53,831, diverse genomes from the NHLBI TOPMed program. Nature 2021; 590:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics 2015;31:782–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 2016;48:1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang J, Howie B, McCarthy S, Memari Y, Walter K, Min JL, Danecek P, Malerba G, Trabetti E, Zheng HF, UK10K Consortium, Gambaro G, Richards JB, Durbin R, Timpson NJ, Marchini J, Soranzo N. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun 2015;6:8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ANthropometric Traits (GIANT) Consortium; DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) Consortium, Madden PAF, Heath AC, Martin NG, Montgomery GW, Weedon MN, Loos RJ, Frayling TM, McCarthy MI, Hirschhorn JN, Goddard ME, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet 2012;44:369–375. S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, ReproGen Consortium; Psychiatric Genomics Consortium; Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium 3, Duncan L, Perry JR, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM. An atlas of genetic correlations across human diseases and traits. Nat Genet 2015;47:1236–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cao J, O’Day DR, Pliner HA, Kingsley PD, Deng M, Daza RM, Zager MA, Aldinger KA, Blecher-Gonen R, Zhang F, Spielmann M, Palis J, Doherty D, Steemers FJ, Glass IA, Trapnell C, Shendure J. A human cell atlas of fetal gene expression. Science 2020;370:eaba7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Watanabe K, Umicevic Mirkov M, de Leeuw CA, van den Heuvel MP, Posthuma D. Genetic mapping of cell type specificity for complex traits. Nat Commun 2019;10:3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sahara M, Santoro F, Sohlmér J, Zhou C, Witman N, Leung CY, Mononen M, Bylund K, Gruber P, Chien KR. Population and single-cell analysis of human cardiogenesis reveals unique LGR5 ventricular progenitors in embryonic outflow tract. Dev Cell 2019;48:475–490.e477. [DOI] [PubMed] [Google Scholar]
- 31. Lahm H, Jia M, Dressen M, Wirth F, Puluca N, Gilsbach R, Keavney BD, Cleuziou J, Beck N, Bondareva O, Dzilic E, Burri M, Konig KC, Ziegelmuller JA, Abou-Ajram C, Neb I, Zhang Z, Doppler SA, Mastantuono E, Lichtner P, Eckstein G, Horer J, Ewert P, Priest JR, Hein L, Lange R, Meitinger T, Cordell HJ, Muller-Myhsok B, Krane M. Congenital heart disease risk loci identified by genome-wide association study in European patients. J Clin Invest 2021;131:e141837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Gruning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018; 46:W537–W544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- 35. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH. Massively parallel digital transcriptional profiling of single cells. Nat Commun 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hao Y, Hao S, Andersen-Nissen E, Mauck WM III, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, Hoffman P, Stoeckius M, Papalexi E, Mimitou EP, Jain J, Srivastava A, Stuart T, Fleming LM, Yeung B, Rogers AJ, McElrath JM, Blish CA, Gottardo R, Smibert P, Satija R. Integrated analysis of multimodal single-cell data. Cell 2021;184:3573–3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, Hao Y, Stoeckius M, Smibert P, Satija R. Comprehensive integration of single-cell data. Cell 2019;177:1888–1902.e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin SW, Beis D, Mitchell T, Chen JN, Stainier DYR. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 2005;132:5199–5209. [DOI] [PubMed] [Google Scholar]
- 40. Westerfield M, Doerry E, Kirkpatrick AE, Driever W, Douglas SA. An on-line database for zebrafish development and genetics research. Semin Cell Dev Biol 1997;8:477–488. [DOI] [PubMed] [Google Scholar]
- 41. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O'Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database Consortium; Neale BM, Daly MJ, MacArthur DG. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020; 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fontana F, Haack T, Reichenbach M, Knaus P, Puceat M, Abdelilah-Seyfried S. Antagonistic activities of Vegfr3/Flt4 and Notch1b fine-tune mechanosensitive signaling during zebrafish cardiac valvulogenesis. Cell Rep 2020;32:107883. [DOI] [PubMed] [Google Scholar]
- 43. Renz M, Otten C, Faurobert E, Rudolph F, Zhu Y, Boulday G, Duchene J, Mickoleit M, Dietrich AC, Ramspacher C, Steed E, Manet-Dupé S, Benz A, Hassel D, Vermot J, Huisken J, Tournier-Lasserve E, Felbor U, Sure U, Albiges-Rizo C, Abdelilah-Seyfried S. Regulation of beta1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev Cell 2015;32:181–190. [DOI] [PubMed] [Google Scholar]
- 44. Veerkamp J, Rudolph F, Cseresnyes Z, Priller F, Otten C, Renz M, Schaefer L, Abdelilah-Seyfried S. Unilateral dampening of Bmp activity by nodal generates cardiac left-right asymmetry. Dev Cell 2013;24:660–667. [DOI] [PubMed] [Google Scholar]
- 45. Beis D, Bartman T, Jin SW, Scott IC, D'Amico LA, Ober EA, Verkade H, Frantsve J, Field HA, Wehman A, Baier H, Tallafuss A, Bally-Cuif L, Chen JN, Stainier DY, Jungblut B. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development 2005;132:4193–4204. [DOI] [PubMed] [Google Scholar]
- 46. Pehlivan T, Pober BR, Brueckner M, Garrett S, Slaugh R, Van Rheeden R, Wilson DB, Watson MS, Hing AV. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am J Med Genet 1999;83:201–206. [PubMed] [Google Scholar]
- 47. Kennedy SJ, Teebi AS, Adatia I, Teshima I. Inherited duplication, dup (8) (p23.1p23.1) pat, in a father and daughter with congenital heart defects. Am J Med Genet 2001;104:79–80. [DOI] [PubMed] [Google Scholar]
- 48. Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, Matsuoka R, Cohen JC, Srivastava D. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003;424:443–447. [DOI] [PubMed] [Google Scholar]
- 49. Tomita-Mitchell A, Maslen CL, Morris CD, Garg V, Goldmuntz E. GATA4 sequence variants in patients with congenital heart disease. J Med Genet 2007;44:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang W, Li X, Shen A, Jiao W, Guan X, Li Z. GATA4 mutations in 486 Chinese patients with congenital heart disease. Eur J Med Genet 2008;51:527–535. [DOI] [PubMed] [Google Scholar]
- 51. Wang E, Sun S, Qiao B, Duan W, Huang G, An Y, Xu S, Zheng Y, Su Z, Gu X, Jin L, Wang H. Identification of functional mutations in GATA4 in patients with congenital heart disease. PLoS One 2013;8:e62138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Duchemin AL, Vignes H, Vermot J. Mechanically activated piezo channels modulate outflow tract valve development through the Yap1 and Klf2-Notch signaling axis. Elife 2019;8:e44706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ponnusamy MP, Seshacharyulu P, Lakshmanan I, Vaz AP, Chugh S, Batra SK. Emerging role of mucins in epithelial to mesenchymal transition. Curr Cancer Drug Targets 2013;13:945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun 2017;485:556–562. [DOI] [PubMed] [Google Scholar]
- 55. Kostina AS, Uspensky VЕ, Irtyuga OB, Ignatieva EV, Freylikhman O, Gavriliuk ND, Moiseeva OM, Zhuk S, Tomilin A, Kostareva АА, Malashicheva AB. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim Biophys Acta 2016;1862:733–740. [DOI] [PubMed] [Google Scholar]
- 56. von Gise A, Pu WT. Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 2012;110:1628–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Krainock M, Toubat O, Danopoulos S, Beckham A, Warburton D, Kim R. Epicardial epithelial-to-mesenchymal transition in heart development and disease. J Clin Med 2016;5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Henderson DJ, Eley L, Turner JE, Chaudhry B. Development of the human arterial valves: understanding bicuspid aortic valve. Front Cardiovasc Med 2021;8:802930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dupuis LE, Osinska H, Weinstein MB, Hinton RB, Kern CB. Insufficient versican cleavage and Smad2 phosphorylation results in bicuspid aortic and pulmonary valves. J Mol Cell Cardiol 2013;60:50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Das S, Rachagani S, Sheinin Y, Smith LM, Gurumurthy CB, Roy HK, Batra SK. Mice deficient in Muc4 are resistant to experimental colitis and colitis-associated colorectal cancer. Oncogene 2016;35:2645–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rowson-Hodel AR, Wald JH, Hatakeyama J, O’Neal WK, Stonebraker JR, VanderVorst K, Saldana MJ, Borowsky AD, Sweeney C, Carraway KL III. Membrane Mucin Muc4 promotes blood cell association with tumor cells and mediates efficient metastasis in a mouse model of breast cancer. Oncogene 2018; 37:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sanchez-Soria P, Camenisch TD. ErbB signaling in cardiac development and disease. Semin Cell Dev Biol 2010;21:929–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author (J.S.).