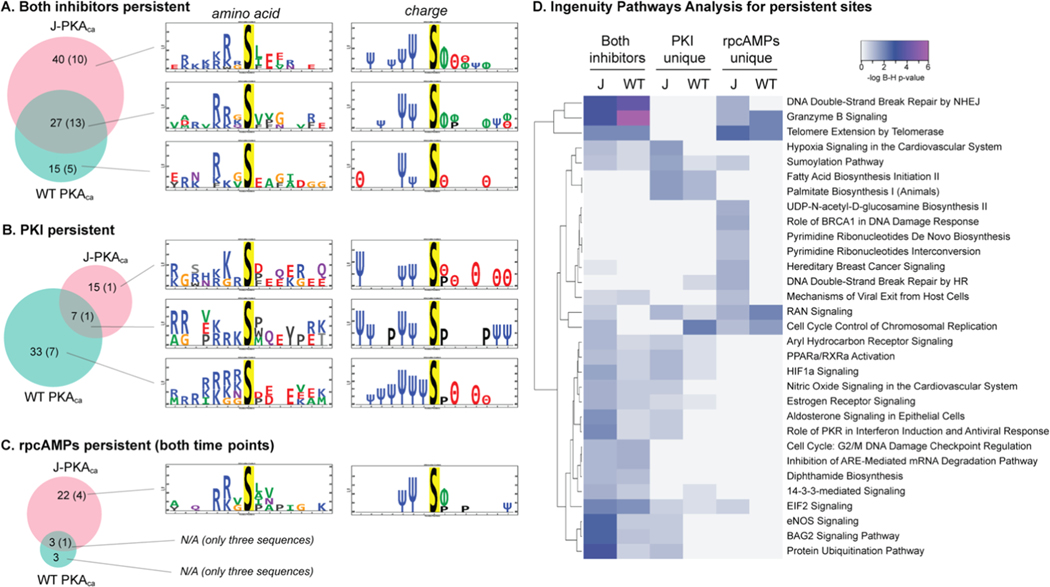
Figure 5.
Motifs observed for persistent phosphosites. Venn diagrams for number of sites that persisted in the presence of inhibitors for each kinase construct (number of those that also had evidence that they may be direct substrates indicated in parentheses) and amino acid/polarity motifs for the sets of sequences associated with each group. (A) Both inhibitors, all time points. (B) Persisting through PKI but not rpcAMPs. (C) Persisting through both time courses of rpcAMPs but not PKI (Venn diagrams created with http://biovenn.nl; motif logos created with PTM-logo23). Charge logo legend: Ψ = basic residue, e.g., K/R; θ = acidic residue, e.g., D/E; ϕ = hydrophobic residue; P = proline. (D) Heatmap generated using https://heatmapper.ca illustrating ingenuity pathways analysis (Supporting Table S4) showing the log10 Benjamini–Hochberg p-values for enrichment of canonical pathways represented by the sites observed as persistent in the presence of both inhibitors or uniquely for each inhibitor, per kinase construct. J = K-PKAca; WT = WT PKAca.