Figure 5.
Clonally related family members of XG005 exhibited striking disparity in neutralizing activity and breadth
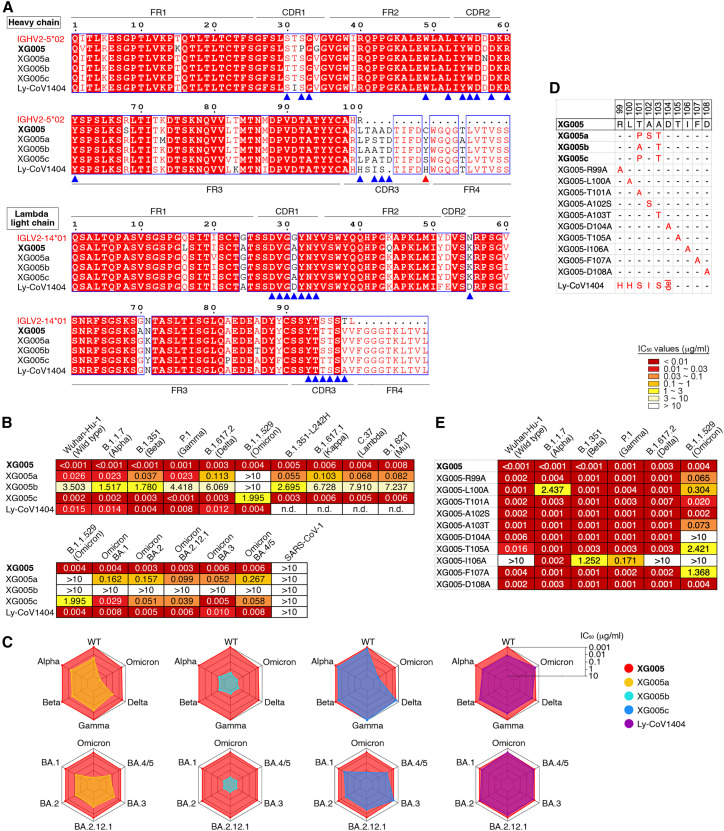
(A) Amino acid sequence alignment for XG005 and its clonally related family members (XG005a, XG005b, and XG005c) and Ly-CoV1404.24,31 IGHV2-5∗02 and IGLV2-14∗01 are the germline reference sequences assigned by IMGT/V-QUEST for IGHV and IGLV, respectively. Boxed red areas are shared among antibodies. The 15 amino acid residues in XG005 IGHV and 14 amino acid residues in XG005 IGLV involved in the recognition of the Omicron RBD are marked by blue arrowheads. The red arrowhead indicates the noncanonical cysteine C109 located in the CDR3 region of XG005 heavy chain.
(B) Pseudovirus neutralization assays. IC50 values for mAbs measured against pseudoviruses of SARS-CoV-1 and SARS-CoV-2 and its variants. Antibodies with IC50 values above 10 μg/mL were shown as >10 μg/mL. The abbreviation n.d. indicates “not determined.” Mean of two independent experiments.
(C) Spider charts for IC50 values of XG005, its three family members, and Ly-CoV1404, which was also encoded by IGHV2-5/IGLV2-14 as XG005.
(D) CDRH3 amino acid sequences of a series of XG005 mutants. The XG005 CDRH3 amino acid sequence is shown completely, while for other mAbs, only the different amino acid residues are labeled in red. The abbreviation del means “amino acid deletion,” while the “-” represents the same amino acid with XG005 CDRH3.
(E) Pseudovirus neutralization assays using XG005 mutants. IC50 values for a series of XG005 mutants against pseudoviruses of SARS-CoV-2 and its variants. Mean of two independent experiments.
See also Figures S5 and S6.