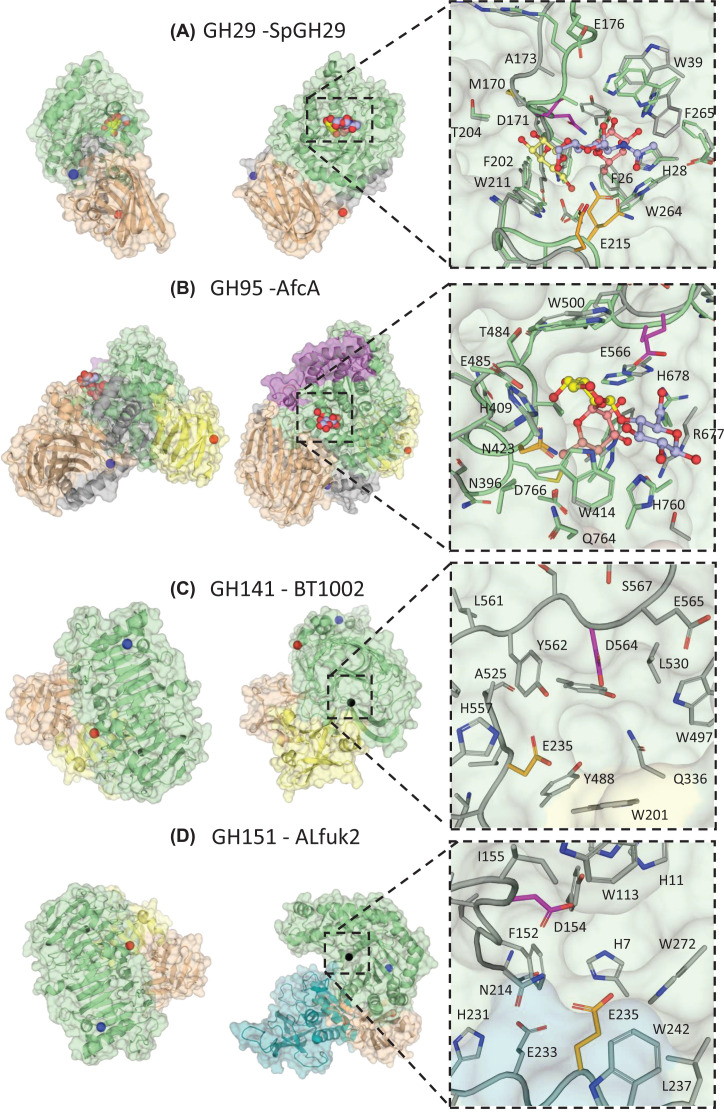
Figure 2. Crystal structures of microbial α-l-fucosidases from different GH families with close up of active sites.
Catalytic modules are shown in green and β-sandwich domains that may have carbohydrate binding properties in light brown and yellow. Catalytic nucleophile residues are coloured magenta and catalytic acid/base residues are coloured in orange. Where possible WT apo crystal structures (grey) have been aligned to their corresponding inactive mutant crystal structures (green) to highlight residue movements upon binding to a substrate like ligand. The N- and C-termini are indicated with blue and red spheres, respectively. Surface representation views are related by a 90° rotation around the y axis. If a substrate complex is not available, the location of the active site is indicated with a black sphere. (A) GH29 fucosidase (SpGH29, apo PDB = 6ORG; D171N; E215Q mutant in complex with LeX PDB = 6ORF). The catalytic domain comprises residues 11-317 and the C terminal β-sandwich module comprises residues 318-451. The bound ligand is shown with Fuc (light red), Gal (yellow) and GlcNAc (light blue). (B) GH95 fucosidase (AfcA, apo PDB = 2EAB; E566A mutant in complex with substrate PDB: 2EAD). The catalytic domain comprises residues 80-133 and 387-778, the N-terminal domain (in light brown) residues 9-79 and 134-293, and the C-terminal β-sandwich module (in yellow) residues 779-896. There is a helical barrel protruding from the N-terminal domain, residues 80-133. The substrate is shown with Gal (yellow), Fuc (light red) and Glc (light blue). C) GH141 fucosidase (BT1002, apo PDB = 5MQP). The catalytic domain comprises residues 1-108 and 296-619, the ancillary β-sandwich domain, residues 109-295 (in yellow for residues 151-251 and in wheat for residues 109-251 and 252-295, according to visual separation into sub domains). D) GH151 fucosidase (ALfuk2, apo PDB = 6TVK). The catalytic domain covers residues 1-336, the C-terminal domain (in wheat), residues 560-660 and the Rossman fold domain (in teal), residues 341-558.