Figure 2. Genetic dissection of Pitx1 regulation reveals the contribution of a pan-limb enhancer (Pen).
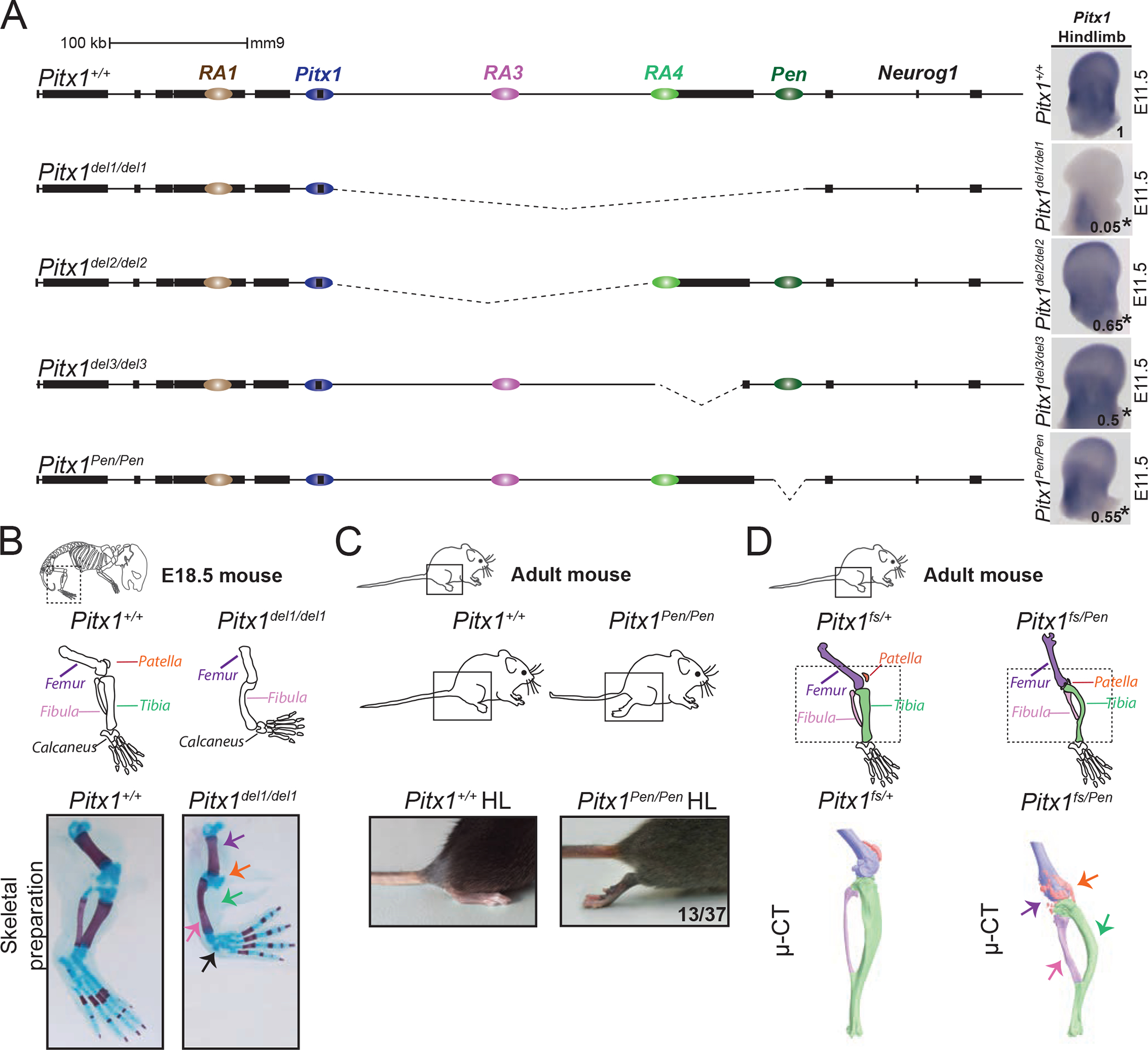
A. CRISPR-Cas9 genetic dissection of the Pitx1 regulatory landscape with mRNA quantification and visualisation of E11.5 hindlimbs using qRT-PCR and WISH. Numbers indicating the average of qRT-PCR fold change compared to wildtype are displayed on the pictures (see figure S2 for quantification). We used a one-sided t-test to evaluate the significance of Pitx1 reduction and an asterisk next to the value indicating a p-value < 0.05. Strong reduction of Pitx1 expression in Pitx1del1/del1 (picture) indicates that its hindlimb regulation is confined to the 330 kb region upstream of the gene (p-value=0.0002, n=4). Pitx1del2/del2 mutants with a 234 kb deletion of the gene desert upstream of Pitx1 show 35% loss of Pitx1 expression (p-value=0.025, n=4). Pitx1del3/del3 mutants carry a 58 kb deletion comprising the H2afy gene and show 50% loss of Pitx1 expression (p-value=0.019, n=3). Pitx1Pen/Pen mice carry a 1.2 kb deletion of the Pen enhancer region and show a 45% loss of Pitx1 expression (p-value=0.012, n=4). B. Pitx1del1/del1 alcian blue and alizarin red skeletal staining at E18.5 demonstrate a Pitx1 loss-of-function phenotype with reduction of the femur, loss of patella and tibia, and small calcaneus (5 skeletal staining out of 5). C. Pitx1Pen/Pen adult mice show a partially penetrant (13 animals out of 37) hindlimb club foot phenotype. D. Pitx1fs/Pen mice display fully penetrant hindlimb malformation (20 animals). μCT of a Pitx1fs/Pen (right) compared to Pitx1fs/+ (middle) adult mice showed a reduced femur (purple), deformed tibia (green) and fibula (pink), as well as a fragmented patella (orange).
