Figure 5. HoxC genes participate in Pitx1 hindlimb regulation.
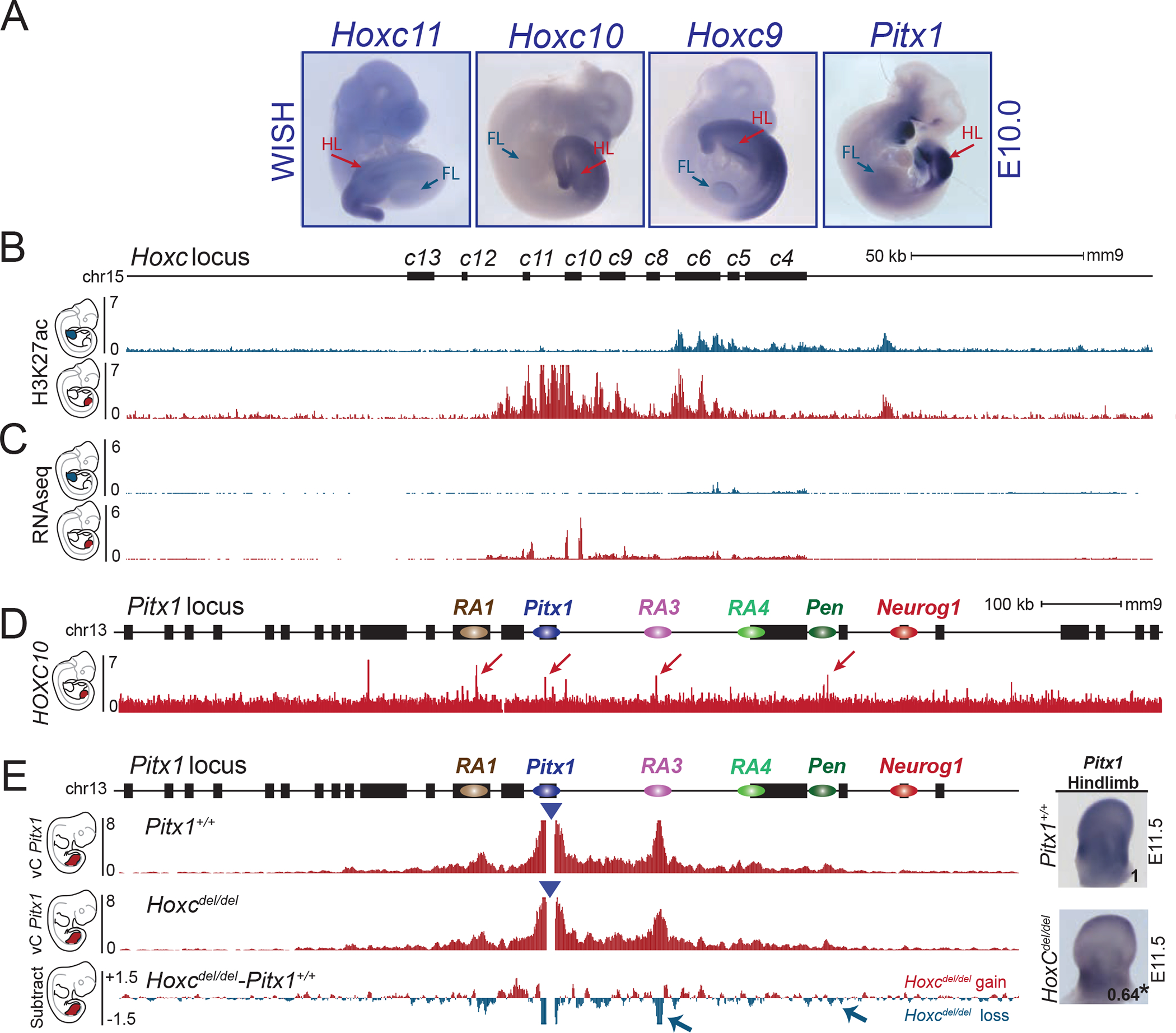
A. WISH of Hoxc11, Hoxc10, Hoxc9 and Pitx1 in E10.5 embryos (WISH were done at least two times per probe). Note the overlapping patterns of expression in lateral plate mesoderm and hindlimbs (HL) and absence of transcript in forelimbs (FL). B. ChIP-seq track for H3K27ac in E10.5 forelimbs (blue) and hindlimbs (red) shows the active state of posterior HoxC genes in hindlimbs and anterior HoxC genes in forelimbs. C. RNA-seq profile (RPKM) of E10.5 forelimb (blue) and hindlimb buds (red) showing expression of posterior HoxC genes in hindlimbs and anterior HoxC genes in forelimbs. D. ChIP-seq track of HOXC10 showing binding at RA1, Pitx1, RA3 and Pen (See red arrows) 25. E. cHiC in Hoxcdel/del hindlimb tissue at E11.5. Derived vC profiles using the Pitx1 viewpoint (blue triangle) shows a significant loss of chromatin interactions between Pitx1 and RA3 as well as Pitx1 and Pen (blue arrow, FDR=0.05). Right: WISH and qRT-PCR of Pitx1 in hindlimb bud shows reduced expression and significant 36% loss of mRNA expression (asterisk: we used a one-sided t-test to evaluate the significance of Pitx1 reduction and found a p-value= 0.02, n= 4 biologically independent wildtype and mutant hindlimb pairs). The number on the picture indicates the average of qRT-PCR fold change compare to wildtype (See figure S7B).
