Figure 6. Ectopic Pitx1-Pen interaction, transcriptional endo-activation and limb malformation induced by chromatin mis-folding.
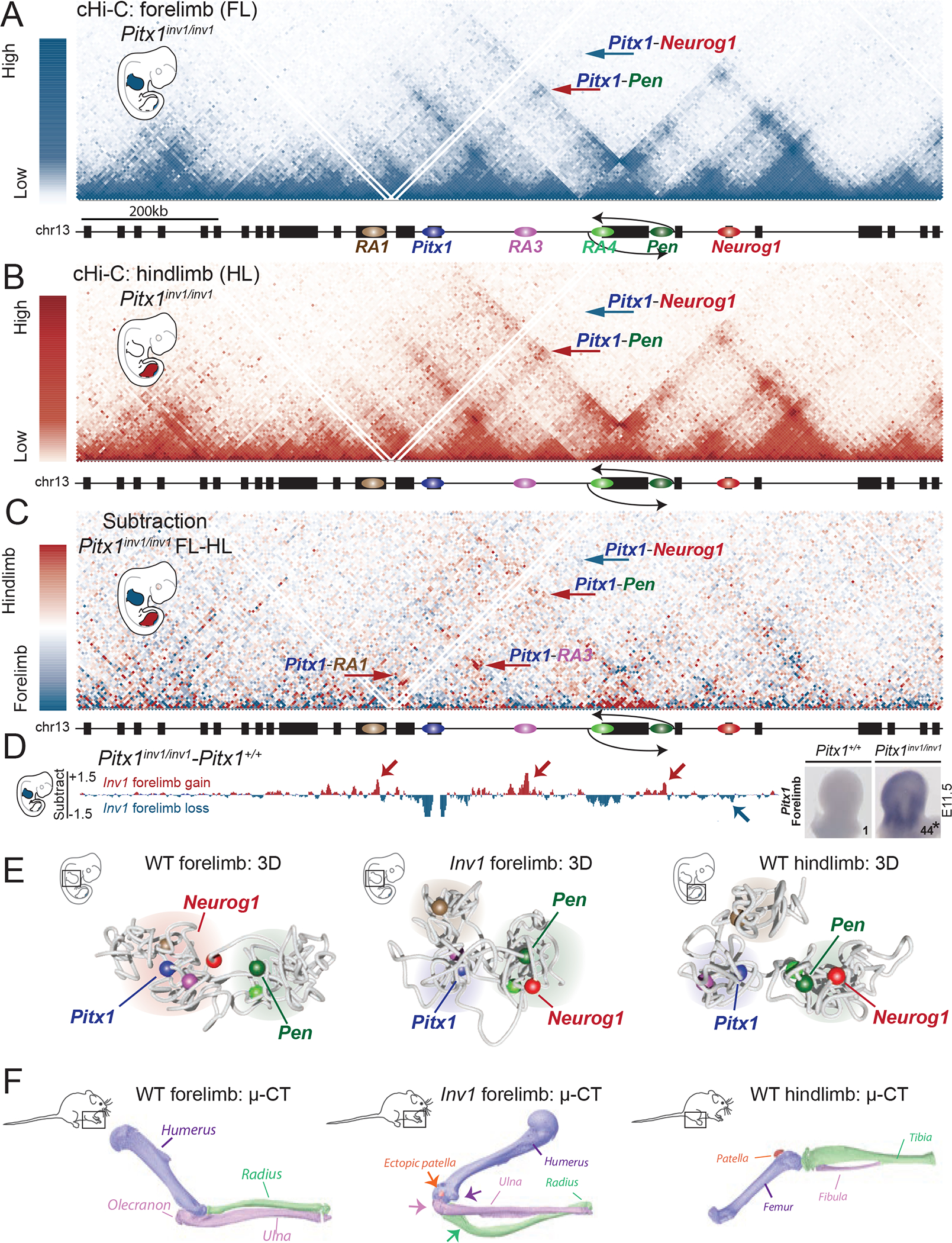
A. cHiC profiles of the Pitx1 locus in Pitx1inv1/inv1 forelimb mapped on wildtype genome. Note the absence of Pitx1-Neurog1 interaction (blue arrow) and the presence of a Pitx1-Pen interaction (red arrow). B. cHiC profiles of the Pitx1 locus in Pitx1inv1/inv1 hindlimbs C. Subtraction maps of Pitx1inv1/inv1 forelimbs and hindlimbs. Note the high similarity of 3D chromatin structure between both tissues in comparison to wild type animals (see Figure 4C). D. Left: subtraction of forelimb wildtpye and inv1/inv1 chromatin interactions show gain of interaction between Pitx1 with RA1, RA3, RA4 and Pen (red arrows) and loss of interaction with Neurog1 (blue arrow) in mutants. Right: WISH and qRT-PCR Pitx1 quantification forelimb tissue demonstrates a strong ectopic expression and 44-fold increase of mRNA expression (asterisk: we used a one-sided t-test to evaluate the significance of Pitx1 increase and found a p-value=1.8×10−7, n=4 biologically independent wildtype and mutant forelimb pairs; see figure S10A). E. 3D model of the Pitx1 locus conformation in wildtype forelimb, Pitx1inv1/inv1 forelimb and wildtype hindlimb. Note that Pitx1inv1/inv1 conformation is hindlimb-like. F. 20 Pitx1inv1/inv1 adult mice display malformed forelimbs and μCT revealed a distorted rotation of the elbow joint (purple arrow), a reduced olecranon (pink arrow), deformed ulna (green arrow), deformed radius as well as an ectopic patella (orange arrow).
