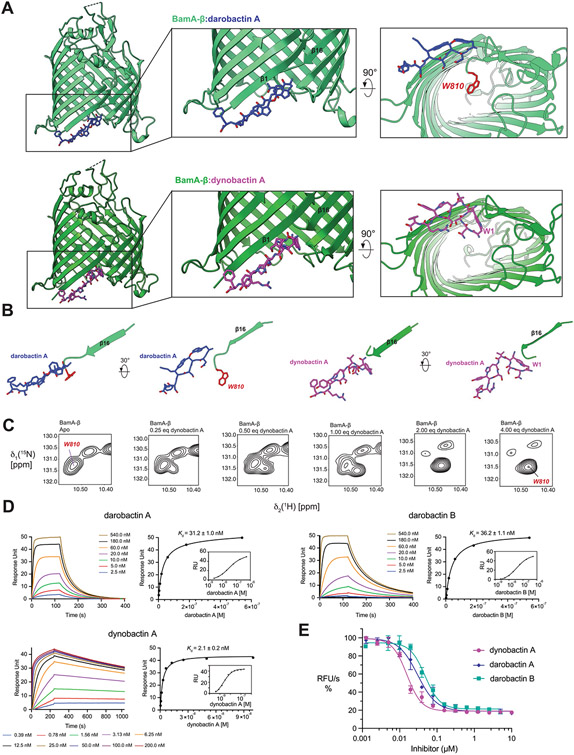
Extended Data Fig. 6 ∣. Unique features of dynobactin A binding.
(A) Comparison of the co-crystal structure of BamA-β with bound darobactin A (PDB:7NRF) and with bound dynobactin A (this work). In the close-up panels, W810 is highlighted in red. (B) Comparison of the orientation of the compound relative to strand β16 of BamA in the two structures. The bulky C-terminal extension of dynobactin A displaces the C-terminus of BamA further into the barrel lumen, with residue W810 becoming flexibly disordered (C) Selected region of a 2D [15N1H]-TROSY spectrum of BamA-β in LDAO micelles upon titration with dynobactin A. Tentative assignment for indole W810 is indicated. (D) Affinity measurements of darobactins and dynobactin A to BamA-β via Surface Plasmon Resonance, sensorgrams and the corresponding steady-state affinity plots show dynobactin A binds one order of magnitude tighter to BamA-barrel than the darobactins. (E) Efficacy of the compounds in inhibiting BAM-mediated folding in native outer membrane vesicles (OMVs) (data are presented as mean values ± SD, n = 2). Fitting of these data resulted in IC50 values of 30 ± 6 nM, 48 ± 8 nM, and 16 ± 2 nM for darobactin A, darobactin B and dynobactin A with a 95% confidence interval, respectively.