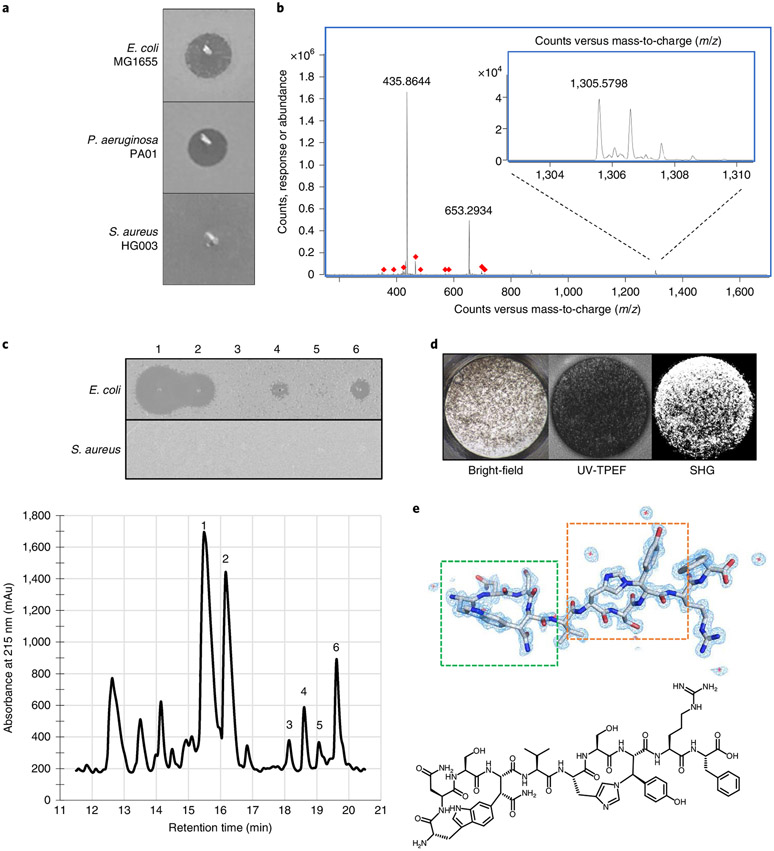
Fig. 3 ∣. Identification of dynobactin A from P. australis.
a, Lawn bio-assay of 20x concentrated P. australis W34 supernatant. b, Mass spectra of dominant Gram-negative selective metabolite, exact mass (M) 1,304.57. c, Bottom: RP-HPLC chromatogram of XAD16N-extracted and SP-FF ion-exchanged P. australis supernatant. Top: isolated peaks assayed against lawns of E. coli and S. aureus. d, Well from high-throughput crystallization screen containing fluorescent crystals. e, Top: cryo-EM microED-generated 3D crystal structure of dynobactin A molecule. Dashed green box, carbon-carbon bond formed between the W1 C6 and the β-carbon of N4; dashed orange box, nitrogen-carbon linkage between the H6 imidazole Nε2 and the β-carbon of Y8. Bottom: accompanying 2D structure.