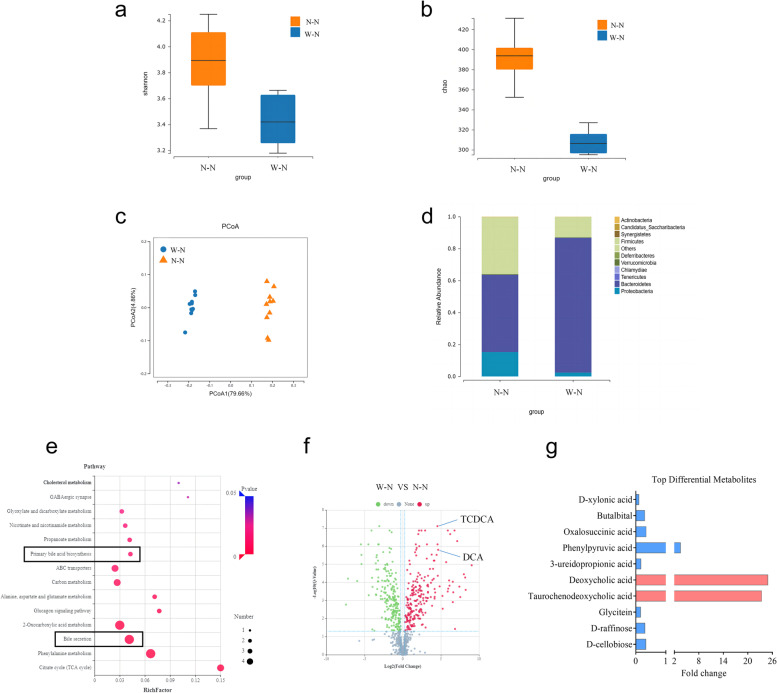
Fig. 3.
MWD induces intestinal microbiota and metabolite shifts in the offspring. Feces from the W-N and N-N group mice were collected for 16s rDNA analysis and untargeted metabolomics analysis. a, b α-diversity analysis of gut microbes reflected by Shannon and Chao indices. c Scatter plots of weighted PCoA for the microbial composition. d Relative abundance of gut microbiota at the phylum level. e KEGG pathway enrichment analyses of the upregulated differential metabolites in the W-N group. The dot size represents the number of differential metabolites, and the dot color represents the corresponding p value. f Scatter plot showing differential metabolites in the W-N group versus the N-N group. Differential metabolites were plotted based on their expression levels (log10 intensity). Red and green dots represented up- and downregulated genes, respectively. Differential bile acids were indicated. g Top 10 differential metabolites in the W-N group versus the N-N group. The y axis shows the name of metabolites, and the x-axis shows the fold change of metabolites