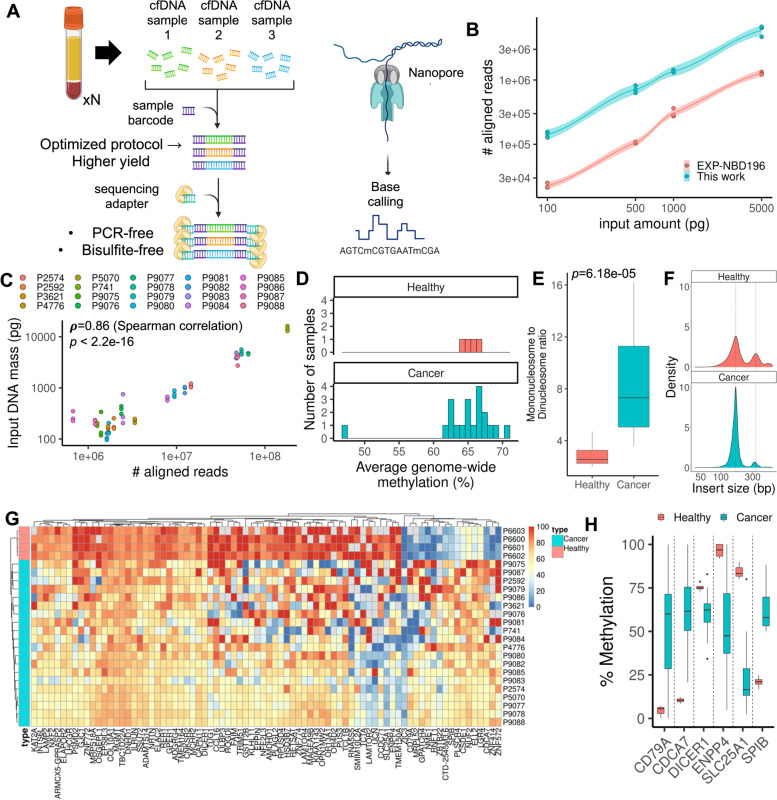
Fig. 1.
Nanopore sequencing of cfDNA. A An optimized protocol for generating cfDNA sequencing libraries enables high-throughput methylation characterization. The key improvement was the optimization of specific end-repair, a-tailing, and ligation conditions to maximize the number of cfDNA fragments available for nanopore sequencing. B Cell-free DNA library comparison. An optimized workflow enables approximately an order of magnitude increase in sequencing yield versus the conventional protocol. C Sequencing yield correlation with input cfDNA. Fluorometric quantification was performed on cancer patient-derived cfDNA samples and compared to the aligned sequencing yield. Each patient is shown as a separate color in triplicate. Correlation and significance value are annotated on the plot. D Genome-wide methylation quantification. The degree of methylation across the genome was computed for healthy and patient-derived cfDNA. E Nucleosome enrichment analysis. The ratio of mononucleosomes to di-nucleosomes was quantified for each tissue type, using a cutoff of 250 bp between mono- and di-nucleosomes. F Distribution of fragment sizes. Example fragment sizes are shown for healthy and patient-derived cfDNA. Mono- and di-nucleosome size peaks are annotated with dotted lines to be 167 bp and 334 bp. G Methylation profiles of healthy- and patient-derived cfDNA. Gene-level methylation values for each sample were determined, and statistically significant ones (q < 0.01) are plotted as a heatmap with the gene-level methylation percentage as the intensities. The heatmap was clustered by gene-level methylation. H Differential methylation. Statistically significant differences in methylation between sample types are shown for several selected genes