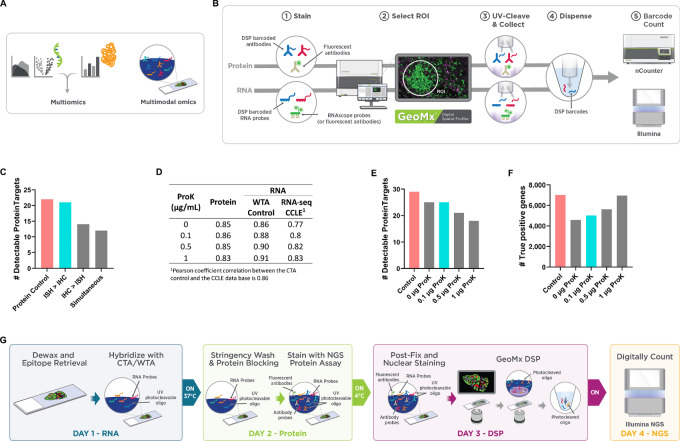
FIGURE 1.
Technical development of the SPG assay. A, Current proteogenomic approaches are multiomic which entails the integration of individual -omic datasets and multimodal omics which involves the simultaneous, codetection of multiple “omes” in a single sample. B, Commercially available GeoMx Assays currently enable high-plex, spatially resolved protein and RNA targets on individual tissue sections with nCounter or NGS quantitative readout. C, Assessment of staining order on the number of protein targets above detection threshold (SNR ≥ 3). FFPE cell line, A431CA, was stained with GeoMx Protein assays (59-plex) for nCounter readout and mock RNA probe (Buffer R only). D, Assessment of varying ProK on the performance of the SPG assay. A 45-CPA was stained with 59-plex GeoMx NGS Protein modules (59-plex) and the GeoMx Whole Transcriptome Atlas (WTA) under proteogenomic and standard assay conditions. E, Plots represent the number of targets above the detection threshold for Pearson correlation on log2-transformed SNR data between the proteogenomic assay and the single-analyte controls along with the CCLE RNA-seq database. Circular ROIs of 200 μm diameter were selected for detailed molecular profiling with the GeoMx DSP. The signal was averaged across replicate AOIs and the SNR was calculated. Protein (SNR ≥ 3; E) and true positives (F) in detectable WTA targets. G, GeoMx SPG workflow enables multimodal omic profiling on a single slide.