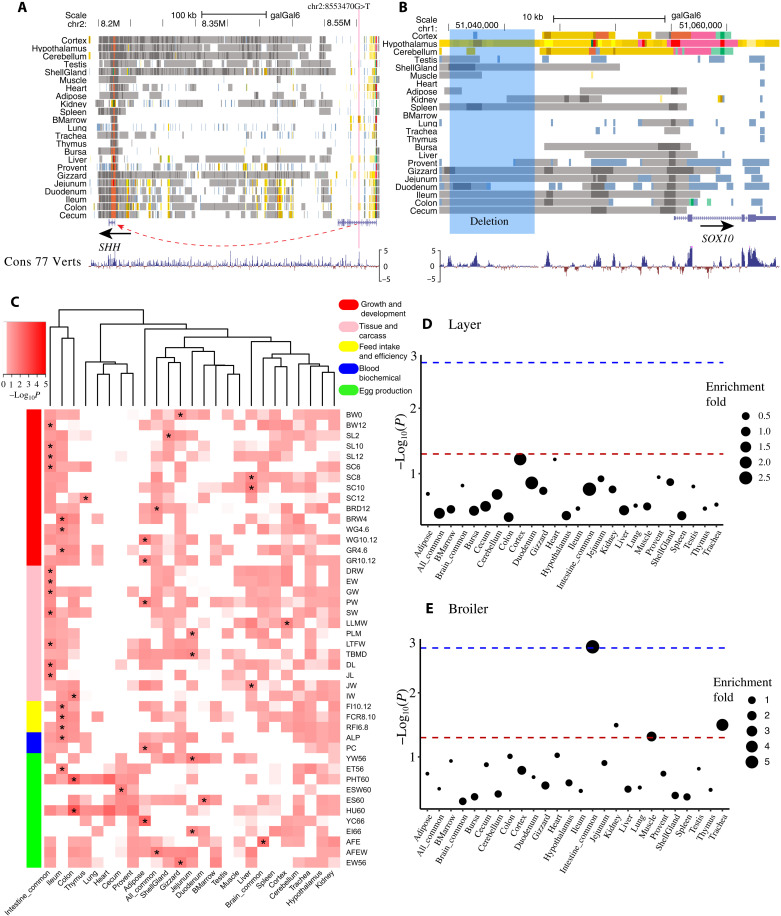
Fig. 5. The regulatory elements involved in chicken monogenic traits, complex traits, and selection signatures.
(A) The predicted chromatin states around a potential causal SNP (chr2:8553470G>T, rs80659072) of polydactyly (chr2:8,546,000-8,561,000) in chickens. The red line indicates the position of the SNP. (B) The predicted chromatin states around a 7.6k deletion (g.51035106_51042744delins, highlighted in blue) at the upstream of SOX10 which causes dark brown/yellow plumage in chickens (chr1:51,034,163-51,064,718). The highlighted region means the deletion region. (C) GWAS enrichment in tissue-specific enhancers (EnhA). “*” indicates that P value is the smallest in this trait, and P < 0.05. (D and E) The enrichment of tissue-specific strong enhancer (EnhA) in selection signatures of layer and broiler, respectively. The dashed lines indicate P < 0.05 (red) and FDR < 0.05 (blue).