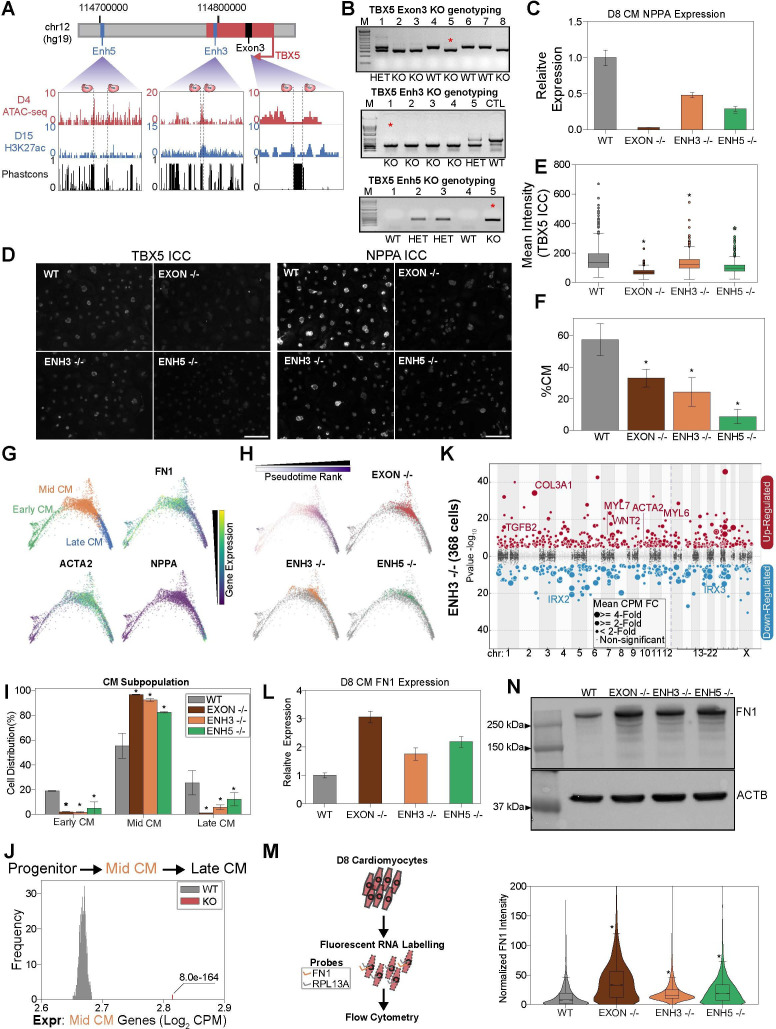
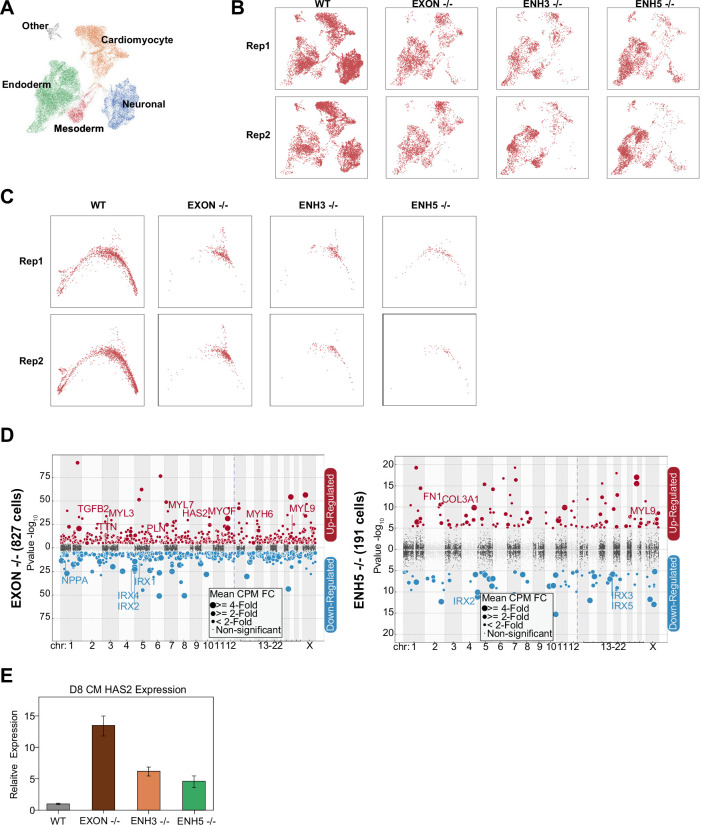
Figure 5. TBX5 enhancer knockouts recapitulate CRISPRi phenotypes.
(A) (Top) TBX5 enhancer knockout strategy. (Bottom) Genome browser snapshots of chromatin status and sequence conservation. Dotted lines denote sgRNA target sites. (B) Genotyping PCR to verify TBX5 knockouts of exon 3 (top), enhancer 3 (middle), and enhancer 5 (bottom). Red asterisk indicates clones used in downstream analysis. (C) NPPA transcript expression in exon and enhancer knockout cells after 8 days of cardiomyocyte (CM) differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with WT cells (n = 3 technical replicates and 2 biological replicates). (D) ICC for TBX5 (left) and NPPA (right) in WT and TBX5 exon 3, enhancer 3, and enhancer 5 knockout cells (Scale bar = 100μm). (E) Quantification of TBX5 ICC (mean intensity) across TBX5 genotyping (*p<0.05 by Mann-Whitney U) (nWT = 2003 cells, nEXON -/- = 780 cells, nENH3 -/- = 1357 cells, nENH5 -/- = 1592 cells). (F) Distribution of WT, TBX5 enhancer KO, or exon KO cells that differentiate into CMs relative to endoderm population (*p<0.05 by hypergeometric test) (nWT = 2 biological replicates, nEXON -/- = 2 biological replicates, nENH3 -/- = 2 biological replicates, nENH5 -/- = 2 biological replicates). (G) (Top left) PHATE visualization of CMs with three Louvain clusters. (Other quadrants) Feature plots of FN1, ACTA2, and NPPA expression. (H) (Top left) Feature plot of pseudotime ranking of CM cells across PHATE trajectory. (Other quadrants) Distribution of TBX5 exon KO and enhancer KO cells across CM trajectory. WT: gray. (I) Distribution of WT, TBX5 exon KO, and enhancer KO cells across three CM subpopulations (*p<0.05 by hypergeometric test) (nWT = 2 biological replicates, nEXON -/- = 2 biological replicates, nENH3 -/- = 2 biological replicates, nENH5 -/- = 2 biological replicates). (J) Averaged expression of mid-CM genes across TBX5 exon and enhancer KO cells (red) and 1000 random samplings of WT mid-CM cells (gray) (*p<0.05 by Z-test). (K) Differentially expressed genes in enhancer 3 KO cells in CM states. Please see description of Manhattan plot in Figure 2I. (L) FN1 transcript expression in exon and enhancer knockout cells after 8 days of CM differentiation. qPCR quantification normalized to reference gene (RPLP0) and then compared with WT cells (n = 3 technical replicates). (M) (Left) Overview of FlowFISH experiment. (Right) Flow cytometry of FN1 RNA FISH intensity normalized by control RPL13A intensity in TBX5 WT and KO lines (*p<0.05 by Mann-Whitney U) (nWT = 4312 cells, nEXON -/- = 4034 cells, nENH3 -/- = 11166 cells, nENH5 -/- = 12286 cells). (N) (Top) FN1 and ACTB (bottom) protein expression in WT, TBX5 exon, and enhancer KO cells after 8 days of CM differentiation.