Figure 1.
CAND1-SCF complexes adopt multiple conformations that collectively reduce CUL1 contacts to SKP1-Fbps and to CAND1
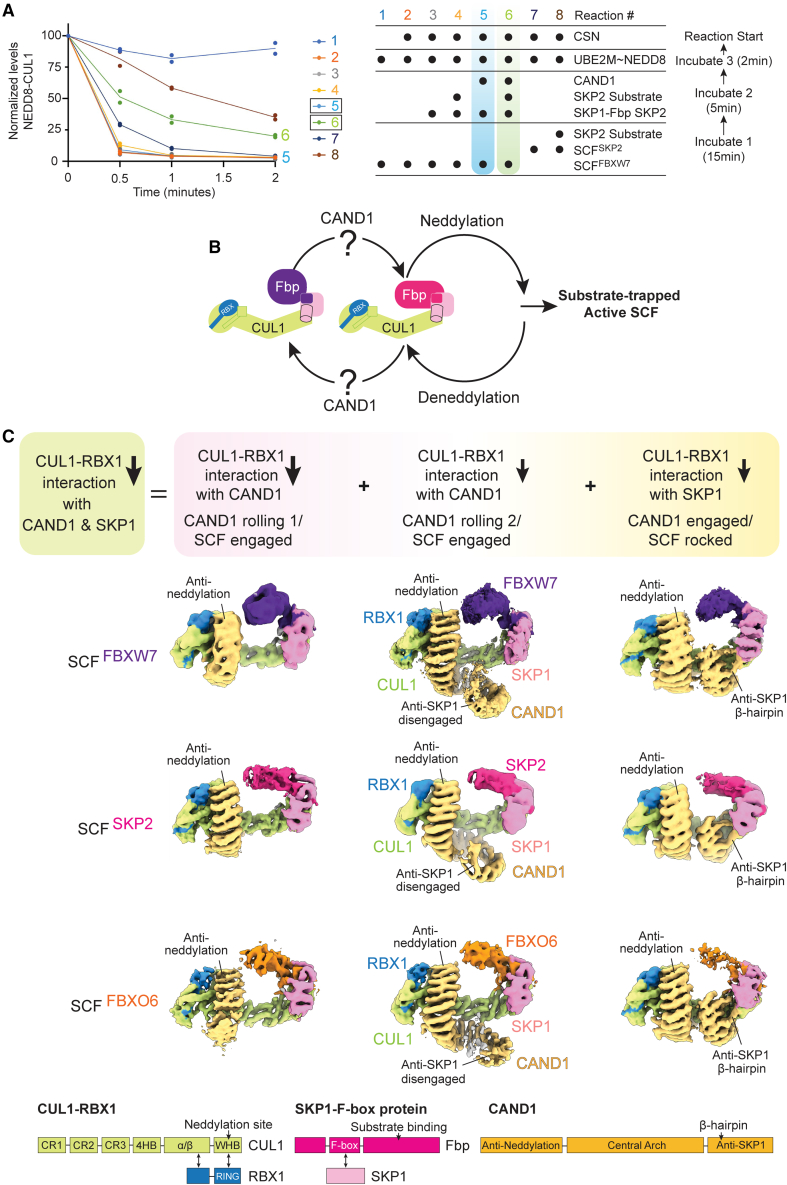
(A) CAND1, neddylation and deneddylation machineries, SKP1-SKP2, and/or the SKP2 substrate complex (phosphorylated p27-CKSHS1-CyclinA-CDK2) were added to a pre-formed SCFFBXW7 or SCFSKP2, as indicated. Systemwide SCF complex formation was read out by CAND1- and SKP2-substrate-dependent retention of NEDD8 linked to CUL1 initially provided in SCFFBXW7. n = 2 dots are plotted.
(B) Cartoon representation of CAND1- and substrate-regulated switching of SKP1-Fbps incorporated into activated SCFs. The structural basis for neddylation, deneddylation, and substrate is well understood.12,27,30,32,35,36,37,48,50 How CAND1 can structurally promote SKP1-Fbp switching in SCFs remains elusive, as indicated by “?”.
(C) Cryo-EM maps showing CAND1-SCF complexes for the Fbps FBXW7, SKP2, and FBXO6. CAND1-SCF complexes form a collection of structures in which CUL1-RBX1 interactions with CAND1 or the SKP1-Fbp are reduced. This would overall lower the barrier to both dissociating: summing relative affinities across individual CAND1-SCF complexes within the ensemble would lower CUL1-RBX1 affinity for both CAND1 and the SKP1-Fbp. Schematics for proteins and their domains are shown below.
See also Figure S1.