Figure 6.
Roles of the CAND1-CUL1-SKP1 interface in SCF disassembly
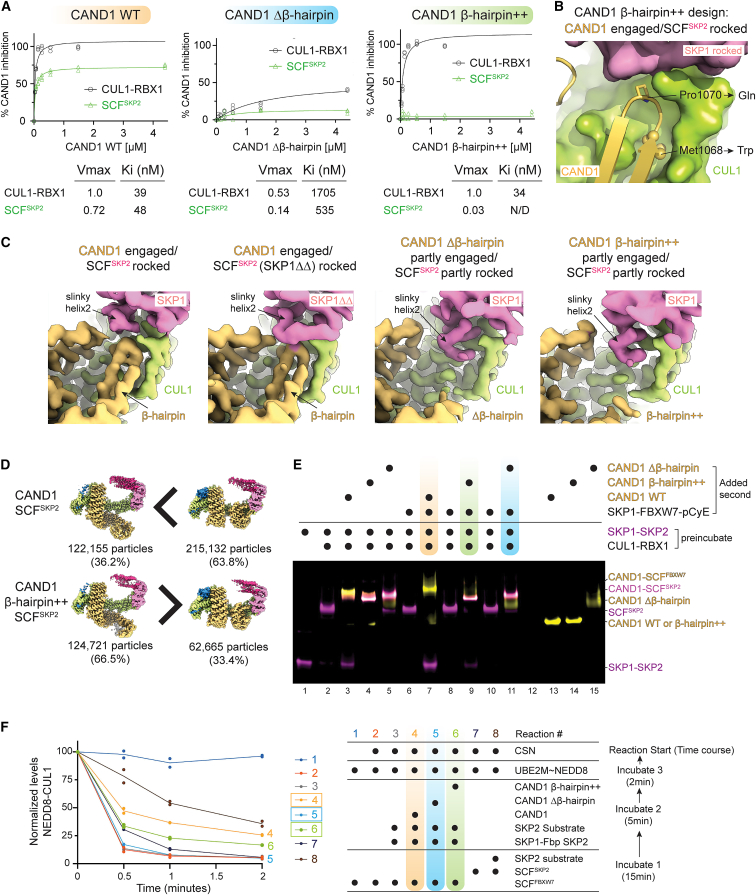
(A) Role of CAND1 mutants in SCF activation, measured by Michaelis-Menten kinetics. Plots show concentration-dependence of indicated CAND1 variant inhibiting neddylation of CUL1 within either CUL1-RBX1 or an SCFSKP2 complex. n = 3 dots are plotted. Rates were normalized to 100 for CUL1-RBX1 without SKP1-Fbp. Kiapp and Vmax values are listed.
(B) Close up of the WT CAND1-CUL1-SKP1 interface, highlighting the sites of the β-hairpin++ mutations (M1068W and P1070Q).
(C) Close ups showing CAND1-CUL1-SKP1 interfaces in cryo-EM maps of indicated complexes.
(D) Compared with the WT complex, cryo-EM data for CAND1β-hairpin++-SCFSKP2 shows relatively more particles with CAND1 in the rolling conformation.
(E) Nondenaturing gel shift assay monitoring SKP1-FBXW7 displacement of SKP1-SKP2 from SCFSKP2, mediated by WT or β-hairpin mutant versions of CAND1. CAND1 is labeled with TAMRA (yellow), SKP2 by Cy5 (magenta) for visualization. FBXW7 samples include Cyclin E phosphopeptide to improve homogeneity of migration.
(F) Effect of CAND1 β-hairpin mutations on assay for CAND1- and substrate-dependent generation and stabilization of an activated SCF in vitro. The assay reflects a CAND1-dependent switch from SCFFBXW7 to SCFSKP2, for which substrate complex protects CUL1 from deneddylation. SKP2 substrate complex is phosphorylated p27-CKSHS1-CyclinA-CDK2. n = 2 dots are plotted.
See also Figure S6.