Figure S7.
Roles of CAND1 across cullin-RING ligase systems, related to Figure 7
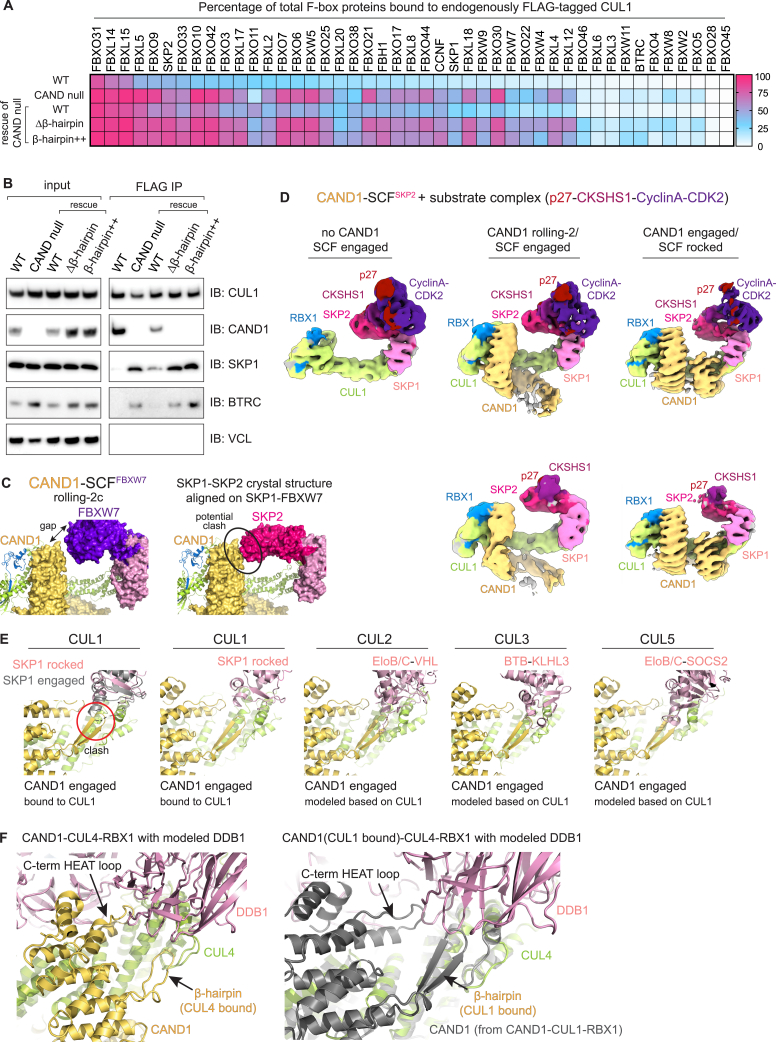
(A) Heatmap showing percentages of FLAG-CUL1-associated versus total Fbps at steady state from WT or CAND-null cells endogenously expressing FLAG-tagged CUL1,9 or the CAND-null cells stably expressing WT CAND1 or the indicated β-hairpin mutants.
(B) Immunoblotting blotting for components of SCFBTRC at steady state, which align with the proteomics results shown in Figure 7B. Chemiluminescent signal with adjusted brightness and contrast is shown.
(C) Docking of SKP1-SKP2 crystal structure46 onto CAND1-SCFFBXW7 rolling-2c conformation shows potential incompatibility between SKP2 and CAND1’s anti-neddylation domain in this orientation.
(D) Cryo-EM structures show addition of substrate complex (phosphorylated p27-CKSHS1-CyclinA-CDK2) alters distribution of CAND1-SCFSKP2 conformations, including SCFSKP2 complexes free of CAND1.
(E) Overlay of various crystal structures of CUL-substrate receptor complexes with the CAND1 engaged/SCFSKP2 rocked structure. Structures were superimposed over CUL CR1 domains. The modeling shows clashing of CAND1’s β-hairpin with CUL2,56 CUL3,62 and CUL563 substrate receptors, suggesting that their disassembly and assembly mechanisms may parallel that described herein for SCFs.
(F) Alignment of CAND1-CUL4-RBX114 (left) and cryo-EM structure of CAND1-CUL1-RBX1 (right) on CUL4-bound DDB1,55 superimposed over the CUL N-terminal CR1 domains, highlighting positions of CAND1’s β-hairpin and C-terminal HEAT repeat loop.