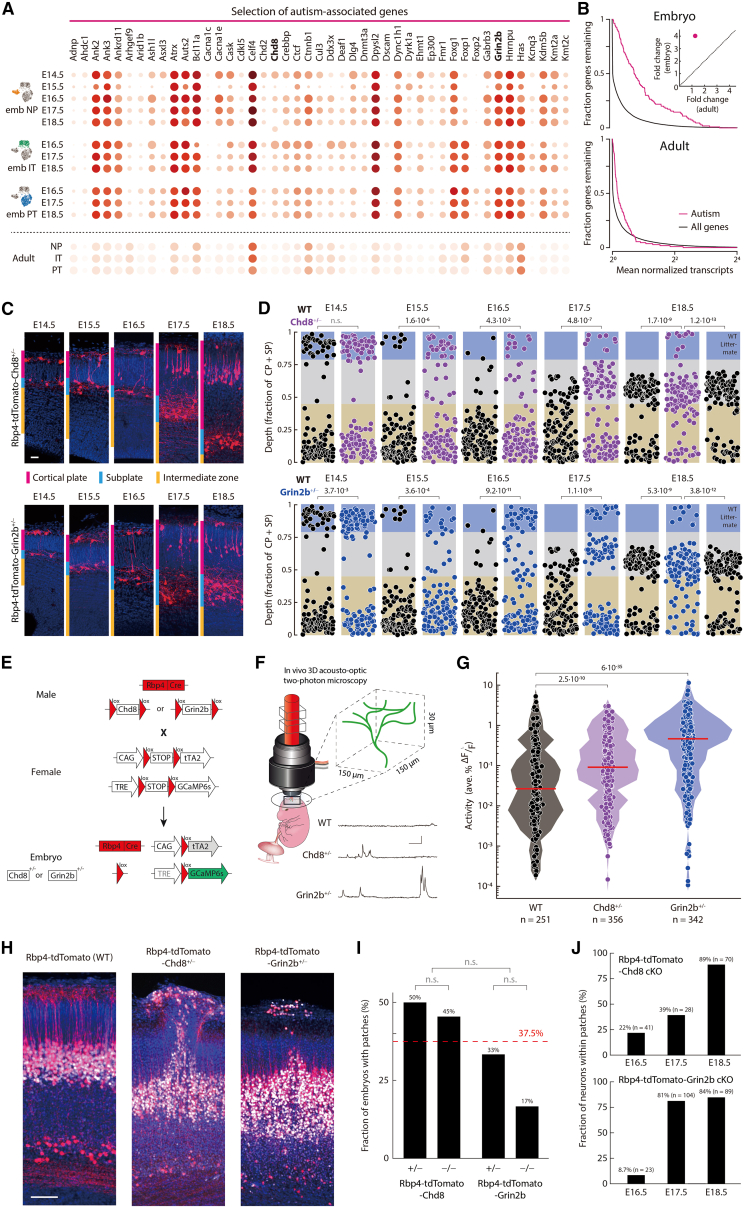
Figure 7.
Perturbing autism-associated genes selectively in Rbp4-Cre neurons disrupts circuit organization and activity during embryonic development
(A) Expression (circles) of selected genes (Data S2) associated with autism spectrum disorder107 in the three Rbp4-Cre neuron types and adult L5-PN types.8 Radius of circles: fraction of cells expressing the gene; color of circles: mean normalized transcripts per cell (log2).
(B) Fraction of genes with a mean transcript count greater than the number of transcripts shown on the x axis, for all genes (black), and genes associated with autism spectrum disorder (magenta) in Rbp4-Cre neurons (top) and adult L5-PNs (bottom). Inset: Fold change of autism-associated gene expression compared to all genes in embryos and adult.
(C) Immunostaining of cortex of Rbp4-tdTomato-Chd8+/− (top) and Rbp4-tdTomato-Grin2b+/− (bottom) mice (Figure S8). Rbp4-Cre neurons (red), Bcl11b, (white), Hoechst (blue).
(D) Normalized depths of Rbp4-Cre neurons (as in Figure 2A) in Rbp4-tdTomato (WT) and the two mutant (top, Chd8+/−; bottom, Grin2b+/−) embryos (Figure S9). 125 neurons from each mouse line, sampled at random. χ2 test.
(E) Mating strategy to generate Rbp4-GCaMP6s-tTA2-Chd8+/− (Chd8+/−) and Rbp4-GCaMP6s-tTA2-Grin2b+/− (Grin2b+/−) embryos.
(F) Example recordings from Rbp4-Cre neurons’ dendrites in Rbp4-GCaMP6s-tTA2 (WT), Grin2b+/−, and Chd8+/− embryos at E16.5 using 3D acousto-optic two-photon microscope.
(G) Distribution of activity in E16.5 embryos, shown in log-scale, for WT and two mutant genotypes. Circles: activity of each neurite; red line: median; shading: distribution. Wilcoxon rank-sum test. n = number of neurites.
(H) Immunostaining of local patches of cortical disorganization in Rbp4-tdTomato-Chd8+/− and Rbp4-tdTomato-Grin2b+/− mice, at E18.5. Rbp4-Cre neurons (red), Bcl11b, (white), Hoechst (blue).
(I) Fraction of mutant mice, of each genotype, showing at least one patch, summed across E16.5 to E18.5. Red line: Average across all four genotypes. Data from 14 (Rbp4-tdTomato-Chd8+/−), 11 (Rbp4-tdTomato-Chd8−/−), 9 (Rbp4-tdTomato-Grin2b+/−), and 6 (Rbp4-tdTomato-Grin2b−/−) embryos. Fisher’s exact test (p = 0.05, prior to Bonferroni correction).
(J) Fraction of neurons within the superficial layer that are located within patches of disorganization, in embryos with at least one patch. n = number of superficial layer neurons on each embryonic day.
Scale bars: 20 μm (C), 25s and 25 %ΔF/F (F), 50 μm (H).
See also Figures S8 and S9.