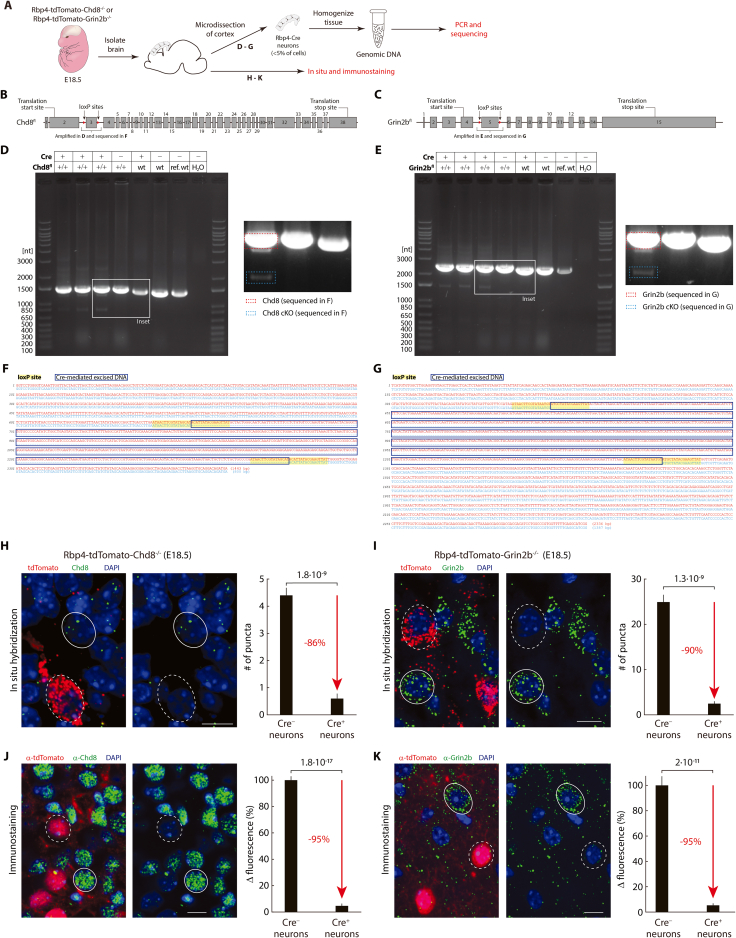
Figure S8.
Crossing floxed Chd8 and Grin2b mouse lines with Rbp4-Cre results in the conditional knockout of Chd8 and Grin2b in Rbp4-Cre neurons, related to Figure 7
(A) Schematic of strategy to demonstrate Cre-mediated excision and resulting selective knockout of Chd8 and Grin2b in E18.5 Rbp4-tdTomato-Chd8−/− or Rbp4-tdTomato-Grin2b−/− embryos.
(B and C) Schematic diagram of Chd8 (B) and Grin2b (C) transcripts (Chd8: Ensembl 212; Grin2b: Ensembl 202), with approximate location of inserted loxP sites, region amplified in D and E (and sequenced in F and G), translational start sites, and translation end sites. Introns not shown to scale.
(D and E) Regions within Chd8 (D) and Grin2b (E) genes, including the loxP sites, were PCR amplified and electrophoretically separated. Above: genotype of embryo for Cre and either floxed Chd8 or Grin2b; ref. wt: commercial mouse genomic DNA. Inset: Zoom in (with increased contrast) showing one example of each genotype and examples of longer (red) and shorter (blue) bands, with relative intensity in agreement with the presence of a small fraction of Cre-expressing cells (i.e. weaker band of the shorter fragment) and a large fraction of cells not expressing Cre (i.e. stronger band of the longer fragment) in the cortical samples.
(F and G) Aligned sequences of excised bands from D and E. Red: sequence of Chd8 (F) and Grin2b (G) bands from longer fragment (derived from cells not expressing Cre) (yellow overlay: loxP sites); blue: sequence of bands from shorter fragment (derived from cells with Cre expression) (blue box: excised region).
(H and I) Left: In situ hybridization of probes against Chd8 (H) or Grin2b (I) (green) and tdTomato (red: marking Rbp4-Cre neurons) in E18.5 Rbp4-tdTomato-Chd8−/− (H) or Rbp4-tdTomato-Grin2b−/− (I) embryos. Example Cre-positive (dashed circles: tdTomato-expressing) and Cre-negative cells (solid circles: lacking tdTomato expression). Right: Number of puncta per cell in Cre-positive compared to Cre-negative cells (mean ± sem). n = 25 cells of each type.
(J and K) Left: Immunostaining against Chd8 (J) or Grin2b (K) (green) and tdTomato (red: marking Rbp4-Cre neurons) in E18.5 Rbp4-tdTomato-Chd8−/− (J) or Rbp4-tdTomato-Grin2b−/− (K) embryos. Example Cre-positive (dashed circles: tdTomato-expressing) and Cre-negative cells (solid circles: lacking tdTomato expression). Right: Quantification of change in fluorescence between Cre-positive cells compared to Cre-negative cells (mean ± sem). Fluorescence was normalized by mean fluorescence within Cre-negative neurons in each slice. n = 50 cells (J) and 30 cells (K) of each type. (H–K) Probability: Wilcoxon rank-sum test. Scale bars: 10 μm (H–K).