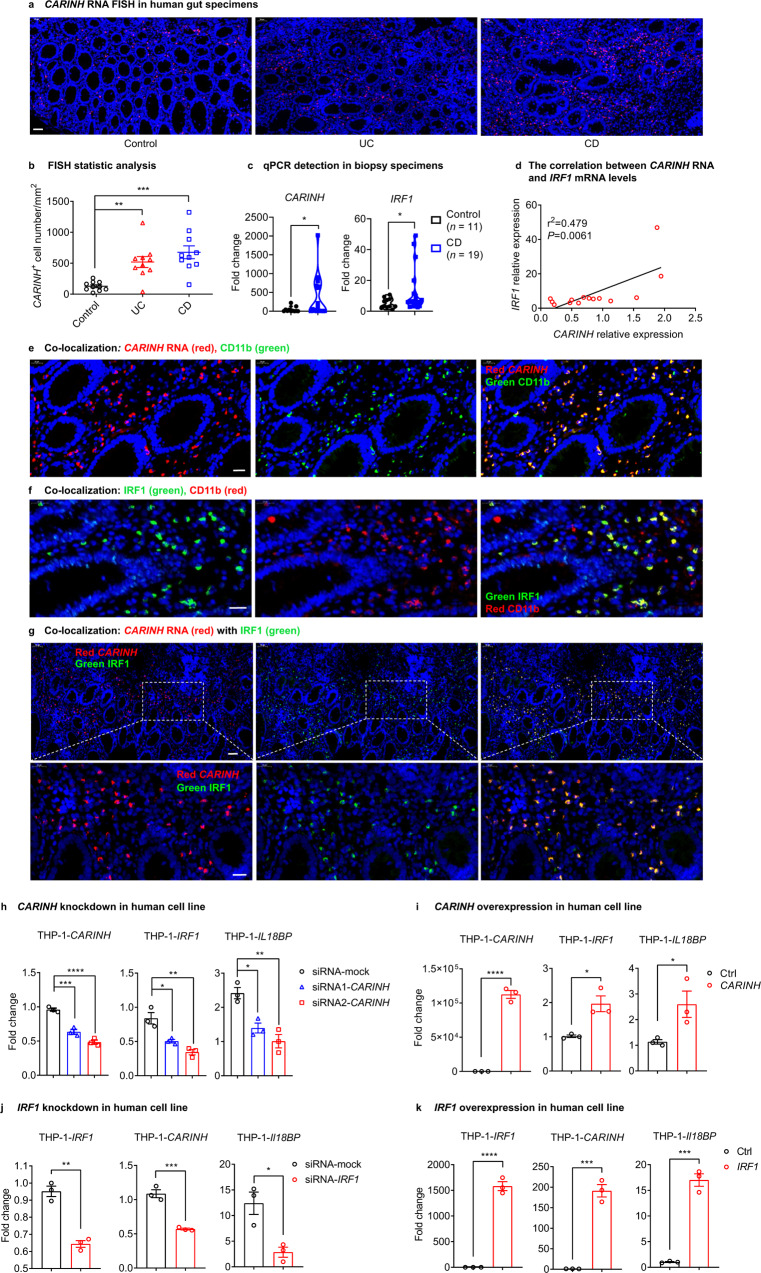
Fig. 5. CARINH is functionally conserved as a regulator of inflammation in human.
a CARINH RNA expression in human gut specimens from control individuals (left, n = 10), UC patients (middle, n = 10), and CD patients (right, n = 10), assessed by RNA FISH. Representative pictures are shown. Scale bar, 50 μm. b Quantification of the FISH results from a, shown as the CARINH RNA-positive cell count per mm2. n = 10 per group. c CARINH and IRF1 mRNA levels are detected by qPCR in terminal ileum biopsy specimens. Control, n = 11; CD, n = 19. d CARINH and IRF1 mRNA levels are detected by qPCR in PBMCs of IBD patients. The correlation was analyzed between CARINH RNA and IRF1 mRNA levels. e FISH and immunofluorescence staining, showing the co-localization of CARINH RNA (red) with CD11b (green). Yellow indicates co-localization. DAPI (blue) stains cell nuclei. Scale bars, 20 μm. f Immunofluorescence staining showing the co-localization of IRF1 (green) with CD11b (red). Yellow indicates co-localization. DAPI (blue) stains cell nuclei. Scale bars, 20 μm. g FISH and immunofluorescent staining showing the co-localization of CARINH RNA (red) with IRF1 (green). Yellow indicates co-localization. DAPI (blue) stains the nuclear of cell. The bottom panel of images are magnified views of the areas indicated by white line boxes above. Up panel scale bar, 50 μm. Bottom panel scale bar, 20 μm. h, i qPCR analyses of human CARINH, IRF1 and IL18BP mRNA expression in THP-1 cells transduced with human CARINH-specific siRNAs or scramble control (h), and with human CARINH-expressing vector or empty vector (i). n = 3 per group. j, k qPCR analyses of human IRF1, CARINH and IL18BP mRNA expression in THP-1 cells transduced with human IRF1-specific siRNAs or scramble control (j), and with human IRF1-expressing vector or empty vector (k). n = 3 per group. The data in h–k are representative of at least three independent experiments. Data represent means ± SEM. Data in b and h were analyzed by one-way ANOVA. Unpaired two-tailed Student’s t-tests were used for other statistical analyses. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.